
Orcun Goksel
Professor
Dept. Information Technology, Uppsala University, Sweden
orcun.goksel@it.uu.se +46 18 471 3460
Office: Room 104147, Ångstrom building 10, Polacksbacken campus, Lägerhyddsvägen 1, 75105 Uppsala, Sweden
Post: Box 337, 75105 Uppsala, Sweden
Head of Computer-assisted Applications in Medicine (CAiM) Group
Dr. Goksel received two BSc degrees in electrical engineering (2001) and in computer science (2002) from Middle East Technical University, Ankara, Turkey. He received his MASc (2004) and PhD (2009) degrees in Electrical and Computer Engineering at the University of British Columbia, Vancouver, Canada. In 2014, he was appointed as an SNSF assistant professor at the Department of Information Technology and Electrical Engineering at ETH Zurich, Switzerland; where he founded the Computer-assisted Applications in Medicine (CAiM) group, which he has been leading. . In 2020, he joined the Department of Information Technology at Uppsala University, Sweden, where he is currently a professor in computerized image processing. He is affiliated with the Centre for Image Analysis as well as the Medtech Science and Innovation Centre. Dr. Goksel has received the 2016 ETH Spark Award (for most promising invention of the year), the 2014 CTI Swiss MedTech Award, and the 2011 WAGS Innovation in Technology Award (for best dissertation in western North America).
- Professor, Department of Information Technology, Uppsala University (2024- )
- Associate Professor, Department of Information Technology, Uppsala University (2020-2024)
- SNSF Professor, D-ITET, ETH Zurich (2014-2022)
- Senior post-doc in Medical Imaging Group at Computer Vision Lab, D-ITET, ETH Zurich (2011-2014)
- Post-doctoral Fellow at Robotics and Control Lab, ECE, UBC (2010)
- Ph.D., Electrical and Computer Eng., UBC (2005-2009)
- M.A.Sc. (Master of Applied Science), Electrical and Computer Eng., UBC (2002-2004)
- B.S., CENG, METU (1998-2002)
- B.S., EEE, METU (1997-2001)
Group CAiM
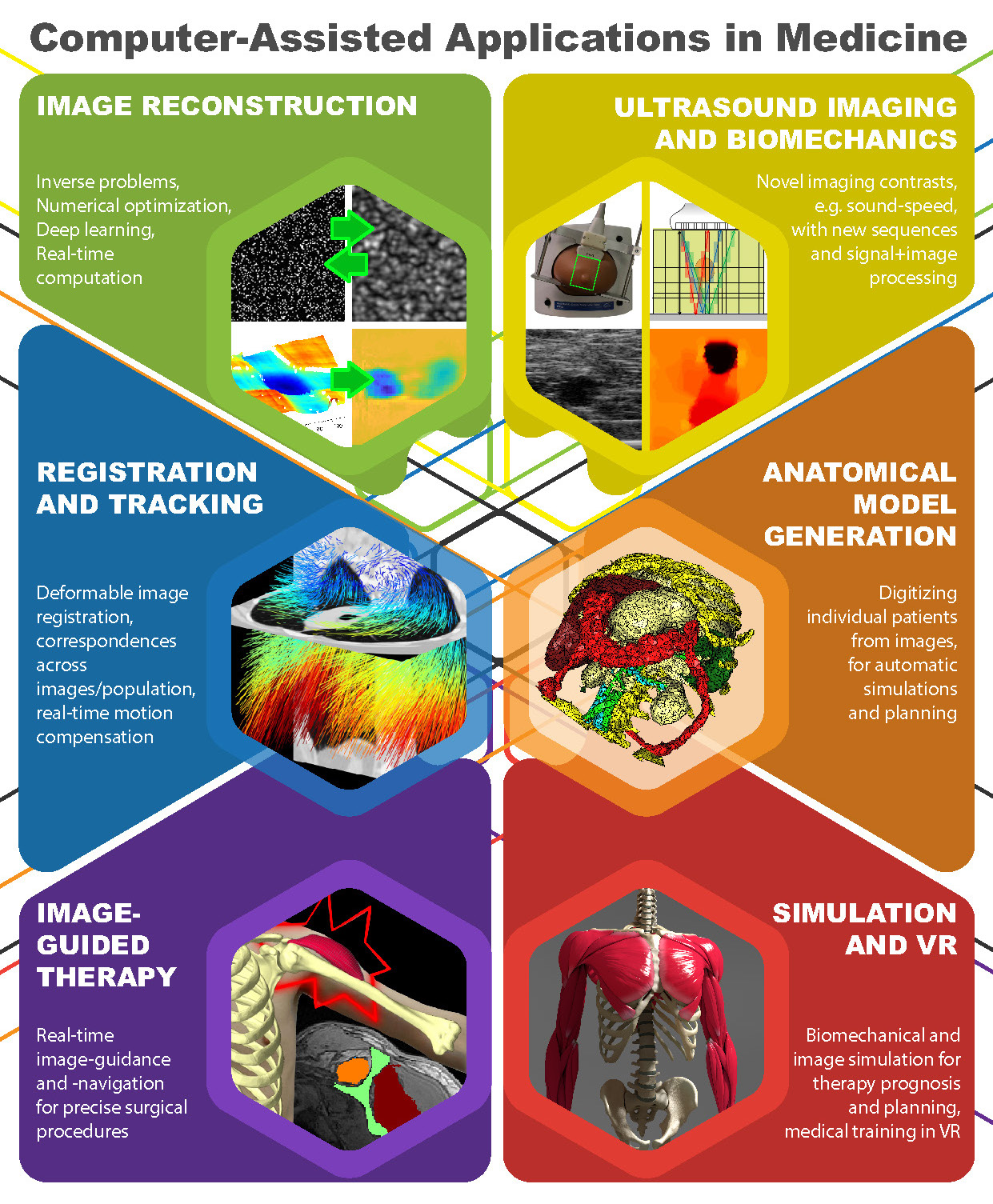
Visual summary of research interests
Publications
Journal Articles:
 [2024] Can Deniz Bezek, Maxim Haas, Richard Rau, and Orcun Goksel:
"Learning the Imaging Model of Speed-of-Sound Reconstruction via a Convolutional Formulation",
IEEE Transactions on Medical Imaging, 2024.
[2024] Can Deniz Bezek, Maxim Haas, Richard Rau, and Orcun Goksel:
"Learning the Imaging Model of Speed-of-Sound Reconstruction via a Convolutional Formulation",
IEEE Transactions on Medical Imaging, 2024.
@article{Bezek_learning_24,
author = {Can Deniz Bezek and Maxim Haas and Richard Rau and Orcun Goksel},
title = {Learning the Imaging Model of Speed-of-Sound Reconstruction via a Convolutional Formulation},
journal = {IEEE Transactions on Medical Imaging},
year = {2024},
doi = {10.1109/TMI.2024.3480690}
}
 [2024] Kevin Thandiackal, Tiziano Portenier, Andrea Giovannini, Maria Gabrani, and Orcun Goksel:
"Generative Feature-driven Image Replay for Continual Learning",
Image and Vision Computing 150(105187), Oct 2024.
[2024] Kevin Thandiackal, Tiziano Portenier, Andrea Giovannini, Maria Gabrani, and Orcun Goksel:
"Generative Feature-driven Image Replay for Continual Learning",
Image and Vision Computing 150(105187), Oct 2024.
@article{Thandiackal_match_24,
author = {Kevin Thandiackal and Tiziano Portenier and Andrea Giovannini and Maria Gabrani and Orcun Goksel},
title = {Generative Feature-driven Image Replay for Continual Learning},
journal = {Image and Vision Computing},
year = {2024},
volume = {150},
number = {105187},
doi = {10.1016/j.imavis.2024.105187}
}
 [2024] Kevin Thandiackal, Luigi Piccinelli, Rajarsi Gupta, Pushpak Pati, and Orcun Goksel:
"Multi-scale Feature Alignment for Continual Learning of Unlabeled Domains",
IEEE Transactions on Medical Imaging 43(7):2599-2609, Jul 2024.
[2024] Kevin Thandiackal, Luigi Piccinelli, Rajarsi Gupta, Pushpak Pati, and Orcun Goksel:
"Multi-scale Feature Alignment for Continual Learning of Unlabeled Domains",
IEEE Transactions on Medical Imaging 43(7):2599-2609, Jul 2024.
@article{Thandiackal_multi-scale_24,
author = {Kevin Thandiackal and Luigi Piccinelli and Rajarsi Gupta and Pushpak Pati and Orcun Goksel},
title = {Multi-scale Feature Alignment for Continual Learning of Unlabeled Domains},
journal = {IEEE Transactions on Medical Imaging},
year = {2024},
volume = {43},
number = {7},
pages = {2599--2609},
doi = {10.1109/TMI.2024.3368365}
}
 [2024] Lingkai Zhu, Fei Zhou, Bozhi Liu, and Orcun Goksel:
"HDRfeat: A Feature-Rich Network for High Dynamic Range Image Reconstruction",
Pattern Recognition Letters 184:148-154, Jun 2024.
[2024] Lingkai Zhu, Fei Zhou, Bozhi Liu, and Orcun Goksel:
"HDRfeat: A Feature-Rich Network for High Dynamic Range Image Reconstruction",
Pattern Recognition Letters 184:148-154, Jun 2024.
@article{Zhu_hdrfeat_24,
author = {Lingkai Zhu and Fei Zhou and Bozhi Liu and Orcun Goksel},
title = {HDRfeat: A Feature-Rich Network for High Dynamic Range Image Reconstruction},
journal = {Pattern Recognition Letters},
year = {2024},
volume = {184},
pages = {148--154},
doi = {10.1016/j.patrec.2024.06.019}
}
 [2024] Iva Doleckova, Tinka Vidovic, Lenka Jandova, Christine Gretzmeier, Alexander A. Navarini, Michael R. MacArthur, Orcun Goksel, Alexander Nyström, and Collin Y. Ewald:
"Calpain inhibition protects against UVB-induced degradation of dermal-epidermal junction-associated proteins",
Journal of Investigative Dermatology, 2024.
[2024] Iva Doleckova, Tinka Vidovic, Lenka Jandova, Christine Gretzmeier, Alexander A. Navarini, Michael R. MacArthur, Orcun Goksel, Alexander Nyström, and Collin Y. Ewald:
"Calpain inhibition protects against UVB-induced degradation of dermal-epidermal junction-associated proteins",
Journal of Investigative Dermatology, 2024.
@article{Doleckova_calpain_24,
author = {Iva Doleckova and Tinka Vidovic and Lenka Jandova and Christine Gretzmeier and Alexander A. Navarini and Michael R. MacArthur and Orcun Goksel and Alexander Nyström and Collin Y. Ewald},
title = {Calpain inhibition protects against UVB-induced degradation of dermal-epidermal junction-associated proteins},
journal = {Journal of Investigative Dermatology},
year = {2024},
doi = {10.1016/j.jid.2024.02.020}
}
 [2024] Alina C. Teuscher, Cyril Statzer, Anita Goyala, Seraina A. Domenig, Ingmar Schoen, Max Hess, Alexander M. Hofer, Andrea Fossati, Viola Vogel, Orcun Goksel, Ruedi Aebersold, and Collin Y. Ewald:
"Longevity interventions modulate mechanotransduction and extracellular matrix homeostasis in C. Elegans",
Nature Communications 15(276), 2024.
[2024] Alina C. Teuscher, Cyril Statzer, Anita Goyala, Seraina A. Domenig, Ingmar Schoen, Max Hess, Alexander M. Hofer, Andrea Fossati, Viola Vogel, Orcun Goksel, Ruedi Aebersold, and Collin Y. Ewald:
"Longevity interventions modulate mechanotransduction and extracellular matrix homeostasis in C. Elegans",
Nature Communications 15(276), 2024.
@article{Teuscher_longevity_24,
author = {Alina C. Teuscher and Cyril Statzer and Anita Goyala and Seraina A. Domenig and Ingmar Schoen and Max Hess and Alexander M. Hofer and Andrea Fossati and Viola Vogel and Orcun Goksel and Ruedi Aebersold and Collin Y. Ewald},
title = {Longevity interventions modulate mechanotransduction and extracellular matrix homeostasis in C. Elegans},
journal = {Nature Communications},
year = {2024},
volume = {15},
number = {276},
doi = {10.1038/s41467-023-44409-2}
}
2023
 [2023] Pushpak Pati, Guillame Jaume, Z. Ayadi, K. Thandiackal, B. Bozorgtabar, M. Gabrani, and Orcun Goksel:
"Weakly Supervised Joint Whole-Slide Segmentation and Classification in Prostate Cancer",
Medical Image Analysis(102915), 2023.
[2023] Pushpak Pati, Guillame Jaume, Z. Ayadi, K. Thandiackal, B. Bozorgtabar, M. Gabrani, and Orcun Goksel:
"Weakly Supervised Joint Whole-Slide Segmentation and Classification in Prostate Cancer",
Medical Image Analysis(102915), 2023.
@article{Pati_weakly_23,
author = {Pushpak Pati and Guillame Jaume and Z Ayadi and K Thandiackal and B Bozorgtabar and M Gabrani and Orcun Goksel},
title = {Weakly Supervised Joint Whole-Slide Segmentation and Classification in Prostate Cancer},
journal = {Medical Image Analysis},
year = {2023},
number = {102915},
doi = {10.1016/j.media.2023.102915}
}
 [2023] Dieter Schweizer, Richard Rau, Can Deniz Bezek, Rahel A. Kubik-Huch, and Orcun Goksel:
"Robust Imaging of Speed-of-Sound Using Virtual Source Transmission",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 70(10):1308-1318, Oct 2023.
[2023] Dieter Schweizer, Richard Rau, Can Deniz Bezek, Rahel A. Kubik-Huch, and Orcun Goksel:
"Robust Imaging of Speed-of-Sound Using Virtual Source Transmission",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 70(10):1308-1318, Oct 2023.
@article{Schweizer_robust_23,
author = {Dieter Schweizer and Richard Rau and Can Deniz Bezek and Rahel A. Kubik-Huch and Orcun Goksel},
title = {Robust Imaging of Speed-of-Sound Using Virtual Source Transmission},
journal = {IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control},
year = {2023},
volume = {70},
number = {10},
pages = {1308--1318},
doi = {10.1109/TUFFC.2023.3303172}
}
 [2023] Boqi Chen, Kevin Thandiackal, Pushpak Pati, and Orcun Goksel:
"Generative appearance replay for continual unsupervised domain adaptation",
Medical Image Analysis 89(102924), Oct 2023.
[2023] Boqi Chen, Kevin Thandiackal, Pushpak Pati, and Orcun Goksel:
"Generative appearance replay for continual unsupervised domain adaptation",
Medical Image Analysis 89(102924), Oct 2023.
@article{Chen_generative_23,
author = {Boqi Chen and Kevin Thandiackal and Pushpak Pati and Orcun Goksel},
title = {Generative appearance replay for continual unsupervised domain adaptation},
journal = {Medical Image Analysis},
year = {2023},
volume = {89},
number = {102924},
url = {https://arxiv.org/abs/2211.11553},
doi = {10.1016/j.media.2023.102924}
}
 [2023] Can Deniz Bezek and Orcun Goksel:
"Analytical Estimation of Beamforming Speed-of-Sound Using Transmission Geometry",
Ultrasonics 10(860725), Sep 2023.
[2023] Can Deniz Bezek and Orcun Goksel:
"Analytical Estimation of Beamforming Speed-of-Sound Using Transmission Geometry",
Ultrasonics 10(860725), Sep 2023.
@article{Bezek_analytical_23,
author = {Can Deniz Bezek and Orcun Goksel},
title = {Analytical Estimation of Beamforming Speed-of-Sound Using Transmission Geometry},
journal = {Ultrasonics},
year = {2023},
volume = {10},
number = {860725},
url = {https://arxiv.org/abs/2211.11553},
doi = {10.1016/j.ultras.2023.107069}
}
2022
 [2022] Bhaskara Rao Chintada, Richard Rau, and Orcun Goksel:
"Spectral Ultrasound Imaging of Speed-of-Sound and Attenuation Using an Acoustic Mirror",
Frontiers in Physics 10(860725), May 2022.
[2022] Bhaskara Rao Chintada, Richard Rau, and Orcun Goksel:
"Spectral Ultrasound Imaging of Speed-of-Sound and Attenuation Using an Acoustic Mirror",
Frontiers in Physics 10(860725), May 2022.
@article{Chintada_spectral_22,
author = {Bhaskara Rao Chintada and Richard Rau and Orcun Goksel},
title = {Spectral Ultrasound Imaging of Speed-of-Sound and Attenuation Using an Acoustic Mirror},
journal = {Frontiers in Physics},
year = {2022},
volume = {10},
number = {860725},
url = {https://arxiv.org/abs/2201.01435},
doi = {10.3389/fphy.2022.860725}
}
 [2022] Alvaro Gomariz, Tiziano Portenier, César Nombela-Arrieta, and Orcun Goksel:
"Probabilistic Spatial Analysis in Quantitative Microscopy with Uncertainty-Aware Cell Detection using Deep Bayesian Regression",
Science Advances 8(5):eabi8295, Feb 2022.
[2022] Alvaro Gomariz, Tiziano Portenier, César Nombela-Arrieta, and Orcun Goksel:
"Probabilistic Spatial Analysis in Quantitative Microscopy with Uncertainty-Aware Cell Detection using Deep Bayesian Regression",
Science Advances 8(5):eabi8295, Feb 2022.
@article{Gomariz_probabilistic_22,
author = {Alvaro Gomariz and Tiziano Portenier and C\'esar Nombela-Arrieta and Orcun Goksel},
title = {Probabilistic Spatial Analysis in Quantitative Microscopy with Uncertainty-Aware Cell Detection using Deep Bayesian Regression},
journal = {Science Advances},
year = {2022},
volume = {8},
number = {5},
pages = {eabi8295},
url = {https://arxiv.org/abs/2102.11865},
doi = {10.1126/sciadv.abi8295}
}
 [2022] Fabien Péan, Philippe Favre, and Orcun Goksel:
"Computational Analysis of Subscapularis Tears and Pectoralis Major Transfers on Muscular Activity",
Clinical Biomechanics 92(105541), Feb 2022.
[2022] Fabien Péan, Philippe Favre, and Orcun Goksel:
"Computational Analysis of Subscapularis Tears and Pectoralis Major Transfers on Muscular Activity",
Clinical Biomechanics 92(105541), Feb 2022.
@article{Pean_computational_22,
author = {Fabien P\'ean and Philippe Favre and Orcun Goksel},
title = {Computational Analysis of Subscapularis Tears and Pectoralis Major Transfers on Muscular Activity},
journal = {Clinical Biomechanics},
year = {2022},
volume = {92},
number = {105541},
url = {https://arxiv.org/abs/2012.14340},
doi = {10.1016/j.clinbiomech.2021.105541}
}
 [2022] Bhaskara R. Chintada, Richard Rau, and Orcun Goksel:
"Nonlinear Characterization of Tissue Viscoelasticity with Acoustoelastic Attenuation of Shear-Waves",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 69(1):38-53, Jan 2022.
[2022] Bhaskara R. Chintada, Richard Rau, and Orcun Goksel:
"Nonlinear Characterization of Tissue Viscoelasticity with Acoustoelastic Attenuation of Shear-Waves",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 69(1):38-53, Jan 2022.
@article{Chintada_nonlinear_22,
author = {Bhaskara R. Chintada and Richard Rau and Orcun Goksel},
title = {Nonlinear Characterization of Tissue Viscoelasticity with Acoustoelastic Attenuation of Shear-Waves},
journal = {IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control},
year = {2022},
volume = {69},
number = {1},
pages = {38-53},
url = {https://arxiv.org/abs/2002.12908},
doi = {10.1109/TUFFC.2021.3105339}
}
 [2022] Pushpak Pati, Guillaume Jaume, Antonio Foncubierta, Florinda Feroce, Anna Maria Anniciello, Giosuè Scognamiglio, Nadia Brancati, Maryse Fiche, Estelle Dubruc, Daniel Riccio, Maurizio Di Bonito, Giuseppe De Pietro, Gerardo Botti, Jean-Philippe Thiran, Maria Frucci, Orcun Goksel, and Maria Gabrani:
"Hierarchical Graph Representations in Digital Pathology",
Medical Image Analysis 75(102264), Jan 2022.
[2022] Pushpak Pati, Guillaume Jaume, Antonio Foncubierta, Florinda Feroce, Anna Maria Anniciello, Giosuè Scognamiglio, Nadia Brancati, Maryse Fiche, Estelle Dubruc, Daniel Riccio, Maurizio Di Bonito, Giuseppe De Pietro, Gerardo Botti, Jean-Philippe Thiran, Maria Frucci, Orcun Goksel, and Maria Gabrani:
"Hierarchical Graph Representations in Digital Pathology",
Medical Image Analysis 75(102264), Jan 2022.
@article{Pati_hierarchical_22,
author = {Pushpak Pati and Guillaume Jaume and Antonio Foncubierta and Florinda Feroce and Anna Maria Anniciello and Giosu\`e Scognamiglio and Nadia Brancati and Maryse Fiche and Estelle Dubruc and Daniel Riccio and Maurizio Di Bonito and Giuseppe De Pietro and Gerardo Botti and Jean-Philippe Thiran and Maria Frucci and Orcun Goksel and Maria Gabrani},
title = {Hierarchical Graph Representations in Digital Pathology},
journal = {Medical Image Analysis},
year = {2022},
volume = {75},
number = {102264},
url = {https://arxiv.org/abs/2102.11057},
doi = {10.1016/j.media.2021.102264}
}
2021
 [2021] Bhaskara Rao Chintada, Richard Rau, and Orcun Goksel:
"Phase-Aberration Correction in Shear-wave Elastography Imaging Using Local Speed-of-Sound Adaptive Beamforming",
Frontiers in Physics: Medical Physics and Imaging 9(690385), Oct 2021.
[2021] Bhaskara Rao Chintada, Richard Rau, and Orcun Goksel:
"Phase-Aberration Correction in Shear-wave Elastography Imaging Using Local Speed-of-Sound Adaptive Beamforming",
Frontiers in Physics: Medical Physics and Imaging 9(690385), Oct 2021.
@article{Chintada_phase-aberration_21,
author = {Bhaskara Rao Chintada and Richard Rau and Orcun Goksel},
title = {Phase-Aberration Correction in Shear-wave Elastography Imaging Using Local Speed-of-Sound Adaptive Beamforming},
journal = {Frontiers in Physics: Medical Physics and Imaging},
year = {2021},
volume = {9},
number = {690385},
url = {https://arxiv.org/abs/2107.02734},
doi = {10.3389/fphy.2021.690385}
}
 [2021] Alvaro Gomariz, Tiziano Portenier, Patrick M. Helbling, Stephan Isringhausen, Ute Suessbier, César Nombela-Arrieta, and Orcun Goksel:
"Modality Attention and Sampling Enables Deep Learning with Heterogeneous Marker Combinations in Fluorescence Microscopy",
Nature Machine Intelligence 3(9):799-811, Aug 2021.
[2021] Alvaro Gomariz, Tiziano Portenier, Patrick M. Helbling, Stephan Isringhausen, Ute Suessbier, César Nombela-Arrieta, and Orcun Goksel:
"Modality Attention and Sampling Enables Deep Learning with Heterogeneous Marker Combinations in Fluorescence Microscopy",
Nature Machine Intelligence 3(9):799-811, Aug 2021.
@article{Gomariz_modality_21,
author = {Alvaro Gomariz and Tiziano Portenier and Patrick M. Helbling and Stephan Isringhausen and Ute Suessbier and C\'esar Nombela-Arrieta and Orcun Goksel},
title = {Modality Attention and Sampling Enables Deep Learning with Heterogeneous Marker Combinations in Fluorescence Microscopy},
journal = {Nature Machine Intelligence},
year = {2021},
volume = {3},
number = {9},
pages = {799-811},
url = {https://arxiv.org/abs/2008.12380},
doi = {10.1038/s42256-021-00379-y}
}
 [2021] Fabien Péan, Philippe Favre, and Orcun Goksel:
"Influence of Rotator Cuff Integrity on Loading and Kinematics Before and After Reverse Shoulder Arthroplasty",
Journal of Biomechanics 129(110778), 2021.
[2021] Fabien Péan, Philippe Favre, and Orcun Goksel:
"Influence of Rotator Cuff Integrity on Loading and Kinematics Before and After Reverse Shoulder Arthroplasty",
Journal of Biomechanics 129(110778), 2021.
@article{Pean_influence_21,
author = {Fabien P\'ean and Philippe Favre and Orcun Goksel},
title = {Influence of Rotator Cuff Integrity on Loading and Kinematics Before and After Reverse Shoulder Arthroplasty},
journal = {Journal of Biomechanics},
year = {2021},
volume = {129},
number = {110778},
url = {https://arxiv.org/abs/2012.09763},
doi = {10.1016/j.jbiomech.2021.110778}
}
 [2021] Justine Robin, Richard Rau, Berkan Lafci, Aileen Schroeter, Michael Reiss, Xosé-Luís Deán-Ben, Orcun Goksel, and Daniel Razansky:
"Hemodynamic response to sensory stimulation in mice: Comparison between functional ultrasound and optoacoustic imaging",
NeuroImage 237:118111, 2021.
[2021] Justine Robin, Richard Rau, Berkan Lafci, Aileen Schroeter, Michael Reiss, Xosé-Luís Deán-Ben, Orcun Goksel, and Daniel Razansky:
"Hemodynamic response to sensory stimulation in mice: Comparison between functional ultrasound and optoacoustic imaging",
NeuroImage 237:118111, 2021.
@article{Robin_hemodynamic_21,
author = {Justine Robin and Richard Rau and Berkan Lafci and Aileen Schroeter and Michael Reiss and Xos\'e-Lu\'is De\'an-Ben and Orcun Goksel and Daniel Razansky},
title = {Hemodynamic response to sensory stimulation in mice: Comparison between functional ultrasound and optoacoustic imaging},
journal = {NeuroImage},
year = {2021},
volume = {237},
pages = {118111},
doi = {10.1016/j.neuroimage.2021.118111}
}
 [2021] Richard Rau, Dieter Schweizer, Valery Vishnevskiy, and Orcun Goksel:
"Speed-of-Sound Imaging using Diverging Waves",
International Journal of Computer Assisted Radiology and Surgery 16:1201-11, Jun 2021.
[2021] Richard Rau, Dieter Schweizer, Valery Vishnevskiy, and Orcun Goksel:
"Speed-of-Sound Imaging using Diverging Waves",
International Journal of Computer Assisted Radiology and Surgery 16:1201-11, Jun 2021.
@article{Rau_speed-of-sound_21,
author = {Richard Rau and Dieter Schweizer and Valery Vishnevskiy and Orcun Goksel},
title = {Speed-of-Sound Imaging using Diverging Waves},
journal = {International Journal of Computer Assisted Radiology and Surgery},
year = {2021},
volume = {16},
pages = {1201-11},
url = {https://arxiv.org/abs/1910.05935},
doi = {10.1007/s11548-021-02426-w}
}
 [2021] Lin Zhang, Tiziano Portenier, and Orcun Goksel:
"Learning Ultrasound Rendering from Cross-Sectional Model Slices for Simulated Training",
International Journal of Computer Assisted Radiology and Surgery 16:721-730, Apr 2021.
[2021] Lin Zhang, Tiziano Portenier, and Orcun Goksel:
"Learning Ultrasound Rendering from Cross-Sectional Model Slices for Simulated Training",
International Journal of Computer Assisted Radiology and Surgery 16:721-730, Apr 2021.
@article{Zhang_learning_21,
author = {Lin Zhang and Tiziano Portenier and Orcun Goksel},
title = {Learning Ultrasound Rendering from Cross-Sectional Model Slices for Simulated Training},
journal = {International Journal of Computer Assisted Radiology and Surgery},
year = {2021},
volume = {16},
pages = {721-730},
url = {https://arxiv.org/abs/2101.08339},
doi = {10.1007/s11548-021-02349-6}
}
 [2021] Lisa Ruby, Sergio J. Sanabria, Katharina Martini, Thomas Frauenfelder, Gerrolt Nico Jukema, Orcun Goksel, and Marga B. Rominger:
"Quantification of immobilization-induced changes in human calf muscle using speed-of-sound ultrasound: An observational pilot study",
Medicine 100(11), Mar 2021.
[2021] Lisa Ruby, Sergio J. Sanabria, Katharina Martini, Thomas Frauenfelder, Gerrolt Nico Jukema, Orcun Goksel, and Marga B. Rominger:
"Quantification of immobilization-induced changes in human calf muscle using speed-of-sound ultrasound: An observational pilot study",
Medicine 100(11), Mar 2021.
@article{Ruby_quantification_21,
author = {Lisa Ruby and Sergio J Sanabria and Katharina Martini and Thomas Frauenfelder and Gerrolt Nico Jukema and Orcun Goksel and Marga B Rominger},
title = {Quantification of immobilization-induced changes in human calf muscle using speed-of-sound ultrasound: An observational pilot study},
journal = {Medicine},
year = {2021},
volume = {100},
number = {11},
doi = {10.1097/MD.0000000000023576}
}
 [2021] Firat Ozdemir, Zixuan Peng, Philipp Fuernstahl, Christine Tanner, and Orcun Goksel:
"Active Learning for Segmentation Based on Bayesian Sample Queries",
Knowledge-Based Systems 214(106531):1-9, Feb 2021.
[2021] Firat Ozdemir, Zixuan Peng, Philipp Fuernstahl, Christine Tanner, and Orcun Goksel:
"Active Learning for Segmentation Based on Bayesian Sample Queries",
Knowledge-Based Systems 214(106531):1-9, Feb 2021.
@article{Ozdemir_active_21,
author = {Firat Ozdemir and Zixuan Peng and Philipp Fuernstahl and Christine Tanner and Orcun Goksel},
title = {Active Learning for Segmentation Based on Bayesian Sample Queries},
journal = {Knowledge-Based Systems},
year = {2021},
volume = {214},
number = {106531},
pages = {1-9},
url = {https://arxiv.org/abs/1912.10493},
doi = {10.1016/j.knosys.2020.106531}
}
 [2021] Richard Rau, Ozan Unal, Dieter Schweizer, Valery Vishnevskiy, and Orcun Goksel:
"Frequency-Dependent Attenuation Reconstruction with an Acoustic Reflector",
Medical Image Analysis 67(101875):1-9, Jan 2021.
[2021] Richard Rau, Ozan Unal, Dieter Schweizer, Valery Vishnevskiy, and Orcun Goksel:
"Frequency-Dependent Attenuation Reconstruction with an Acoustic Reflector",
Medical Image Analysis 67(101875):1-9, Jan 2021.
* Runner-up for the MICCAI Elsevier MedIA Prize
@article{Rau_frequency-dependent_21,
author = {Richard Rau and Ozan Unal and Dieter Schweizer and Valery Vishnevskiy and Orcun Goksel},
title = {Frequency-Dependent Attenuation Reconstruction with an Acoustic Reflector},
journal = {Medical Image Analysis},
year = {2021},
volume = {67},
number = {101875},
pages = {1-9},
url = {https://arxiv.org/abs/2003.05658},
doi = {10.1016/j.media.2020.101875}
}
 [2021] Pushpak Pati, Antonio Foncubierta-Rodríguez, Orcun Goksel, and Maria Gabrani:
"Reducing Annotation Effort in Digital Pathology: A Co-Representation Learning Framework for Classification Tasks",
Medical Image Analysis 67(101859):1-17, Jan 2021.
[2021] Pushpak Pati, Antonio Foncubierta-Rodríguez, Orcun Goksel, and Maria Gabrani:
"Reducing Annotation Effort in Digital Pathology: A Co-Representation Learning Framework for Classification Tasks",
Medical Image Analysis 67(101859):1-17, Jan 2021.
@article{Pati_reducing_21,
author = {Pushpak Pati and Antonio Foncubierta-Rodr\'{i}guez and Orcun Goksel and Maria Gabrani},
title = {Reducing Annotation Effort in Digital Pathology: A Co-Representation Learning Framework for Classification Tasks},
journal = {Medical Image Analysis},
year = {2021},
volume = {67},
number = {101859},
pages = {1-17},
doi = {10.1016/j.media.2020.101859}
}
2020
 [2020] Lin Zhang, Valery Vishnevskiy, and Orcun Goksel:
"Deep Network for Scatterer Distribution Estimation for Ultrasound Image Simulation",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 67(12):2553-2564, Dec 2020.
[2020] Lin Zhang, Valery Vishnevskiy, and Orcun Goksel:
"Deep Network for Scatterer Distribution Estimation for Ultrasound Image Simulation",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 67(12):2553-2564, Dec 2020.
@article{Zhang_deepN_20,
author = {Lin Zhang and Valery Vishnevskiy and Orcun Goksel},
title = {Deep Network for Scatterer Distribution Estimation for Ultrasound Image Simulation},
journal = {IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control},
year = {2020},
volume = {67},
number = {12},
pages = {2553-2564},
url = {https://arxiv.org/abs/2006.10166},
doi = {10.1109/TUFFC.2020.3018424}
}
 [2020] Melanie Bernhardt, Valery Vishnevskiy, Richard Rau, and Orcun Goksel:
"Training Variational Networks with Multi-Domain Simulations: Speed-of-Sound Image Reconstruction",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 67(12):2584-2594, Dec 2020.
[2020] Melanie Bernhardt, Valery Vishnevskiy, Richard Rau, and Orcun Goksel:
"Training Variational Networks with Multi-Domain Simulations: Speed-of-Sound Image Reconstruction",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 67(12):2584-2594, Dec 2020.
@article{Bernhardt_training_20,
author = {Melanie Bernhardt and Valery Vishnevskiy and Richard Rau and Orcun Goksel},
title = {Training Variational Networks with Multi-Domain Simulations: Speed-of-Sound Image Reconstruction},
journal = {IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control},
year = {2020},
volume = {67},
number = {12},
pages = {2584-2594},
url = {https://arxiv.org/abs/2006.14395},
doi = {10.1109/TUFFC.2020.3010186}
}
 [2020] Fabien Péan and Orcun Goksel:
"Surface-based modeling of muscles: Functional simulation of the shoulder",
Medical Engineering and Physics 82:1-12, 2020.
[2020] Fabien Péan and Orcun Goksel:
"Surface-based modeling of muscles: Functional simulation of the shoulder",
Medical Engineering and Physics 82:1-12, 2020.
@article{Pean_surface-based_20,
author = {Fabien P\'ean and Orcun Goksel},
title = {Surface-based modeling of muscles: Functional simulation of the shoulder},
journal = {Medical Engineering and Physics},
year = {2020},
volume = {82},
pages = {1-12},
doi = {10.1016/j.medengphy.2020.04.010}
}
 [2020] Sabine Kling, Hossein Khodadadi, and Orcun Goksel:
"Optical coherence elastography based corneal strain imaging during low-amplitude intraocular pressure modulation",
Frontiers in Bioengineering and Biotechnology: Biomechanics, Jan 2020.
[2020] Sabine Kling, Hossein Khodadadi, and Orcun Goksel:
"Optical coherence elastography based corneal strain imaging during low-amplitude intraocular pressure modulation",
Frontiers in Bioengineering and Biotechnology: Biomechanics, Jan 2020.
@article{Kling_optical_20,
author = {Sabine Kling and Hossein Khodadadi and Orcun Goksel},
title = {Optical coherence elastography based corneal strain imaging during low-amplitude intraocular pressure modulation},
journal = {Frontiers in Bioengineering and Biotechnology: Biomechanics},
year = {2020},
doi = {10.3389/fbioe.2019.00453}
}
2019
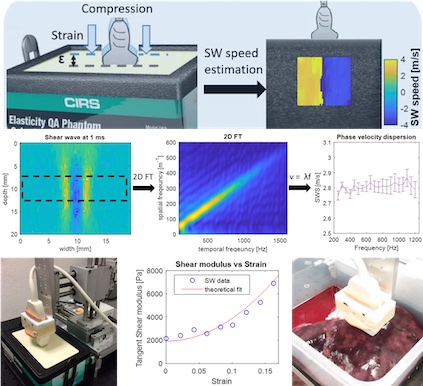 [2019] Corin F. Otesteanu, Bhaskara R. Chintada, Marga B. Rominger, Sergio J. Sanabria, and Orcun Goksel:
"Spectral Quantification of Nonlinear Elasticity using Acoustoelasticity and Shear-Wave Dispersion",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 66(12):1845-1855, Aug 2019.
[2019] Corin F. Otesteanu, Bhaskara R. Chintada, Marga B. Rominger, Sergio J. Sanabria, and Orcun Goksel:
"Spectral Quantification of Nonlinear Elasticity using Acoustoelasticity and Shear-Wave Dispersion",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 66(12):1845-1855, Aug 2019.
@article{Otesteanu_spectral_19,
author = {Corin F Otesteanu and Bhaskara R Chintada and Marga B Rominger and Sergio J Sanabria and Orcun Goksel},
title = {Spectral Quantification of Nonlinear Elasticity using Acoustoelasticity and Shear-Wave Dispersion},
journal = {IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control},
year = {2019},
volume = {66},
number = {12},
pages = {1845-1855},
url = {https://www.research-collection.ethz.ch/bitstream/handle/20.500.11850/362253/Otesteanu_spectral_19pre.pdf},
doi = {10.1109/TUFFC.2019.2933952}
}
 [2019] Rastislav Starkov, Lin Zhang, Michael Bajka, Christine Tanner, and Orcun Goksel:
"Ultrasound Simulation with Deformable and Patient-Specific Scatterer Maps",
Int J Computer Assisted Radiology and Surgery 14(9):1589-1599, Aug 2019.
[2019] Rastislav Starkov, Lin Zhang, Michael Bajka, Christine Tanner, and Orcun Goksel:
"Ultrasound Simulation with Deformable and Patient-Specific Scatterer Maps",
Int J Computer Assisted Radiology and Surgery 14(9):1589-1599, Aug 2019.
@article{Starkov_ultrasound_19d,
author = {Rastislav Starkov and Lin Zhang and Michael Bajka and Christine Tanner and Orcun Goksel},
title = {Ultrasound Simulation with Deformable and Patient-Specific Scatterer Maps},
journal = {Int J Computer Assisted Radiology and Surgery},
year = {2019},
volume = {14},
number = {9},
pages = {1589-1599},
doi = {10.1007/s11548-019-02054-5}
}
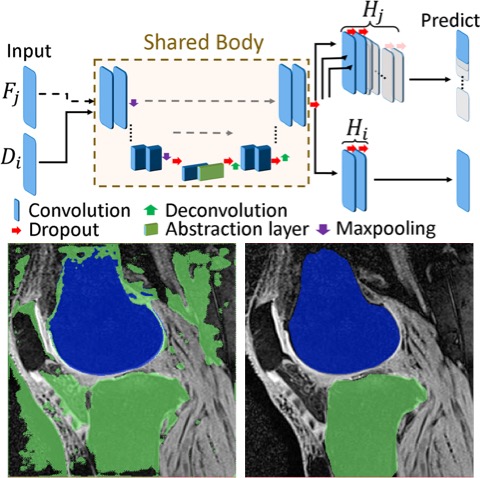 [2019] Firat Ozdemir and Orcun Goksel:
"Extending pretrained segmentation networks with additional anatomical structures",
Int J Computer Assisted Radiology and Surgery 14(7):1187-1195, Jul 2019.
[2019] Firat Ozdemir and Orcun Goksel:
"Extending pretrained segmentation networks with additional anatomical structures",
Int J Computer Assisted Radiology and Surgery 14(7):1187-1195, Jul 2019.
@article{Ozdemir_extending_19,
author = {Firat Ozdemir and Orcun Goksel},
title = {Extending pretrained segmentation networks with additional anatomical structures},
journal = {Int J Computer Assisted Radiology and Surgery},
year = {2019},
volume = {14},
number = {7},
pages = {1187-1195},
url = {https://arxiv.org/abs/1811.04634},
doi = {10.1007/s11548-019-01984-4}
}
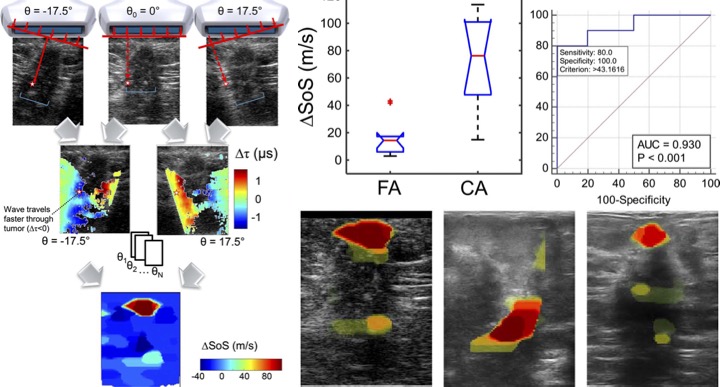 [2019] Lisa Ruby, Sergio J. Sanabria, Katharina Martini, Konstantin J. Dedes, Denise Vorburger, Ece Oezkan, Thomas Frauenfelder, Orcun Goksel, and Marga B. Rominger:
"Breast Cancer Assessment With Pulse-Echo Speed of Sound Ultrasound From Intrinsic Tissue Reflections: Proof-of-Concept",
Investigative Radiology 54(7):419-427, Jul 2019.
[2019] Lisa Ruby, Sergio J. Sanabria, Katharina Martini, Konstantin J. Dedes, Denise Vorburger, Ece Oezkan, Thomas Frauenfelder, Orcun Goksel, and Marga B. Rominger:
"Breast Cancer Assessment With Pulse-Echo Speed of Sound Ultrasound From Intrinsic Tissue Reflections: Proof-of-Concept",
Investigative Radiology 54(7):419-427, Jul 2019.
@article{Ruby_breast_19,
author = {Lisa Ruby and Sergio J. Sanabria and Katharina Martini and Konstantin J. Dedes and Denise Vorburger and Ece Oezkan and Thomas Frauenfelder and Orcun Goksel and Marga B. Rominger},
title = {Breast Cancer Assessment With Pulse-Echo Speed of Sound Ultrasound From Intrinsic Tissue Reflections: Proof-of-Concept},
journal = {Investigative Radiology},
year = {2019},
volume = {54},
number = {7},
pages = {419-427},
url = {https://www.zora.uzh.ch/id/eprint/170532/1/document.pdf},
doi = {10.1097/RLI.0000000000000553}
}
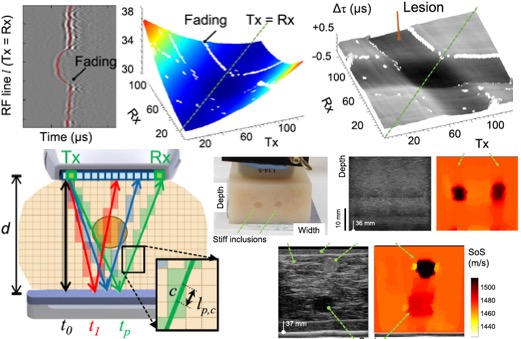 [2019] Sergio J. Sanabria, Marga B. Rominger, and Orcun Goksel:
"Speed-of-Sound Imaging Based on Reflector Delineation",
IEEE Trans Biomedical Engineering 66(7):1949-1962, Jul 2019.
[2019] Sergio J. Sanabria, Marga B. Rominger, and Orcun Goksel:
"Speed-of-Sound Imaging Based on Reflector Delineation",
IEEE Trans Biomedical Engineering 66(7):1949-1962, Jul 2019.
@article{Sanabria_speed-of-sound_19,
author = {Sergio J Sanabria and Marga B Rominger and Orcun Goksel},
title = {Speed-of-Sound Imaging Based on Reflector Delineation},
journal = {IEEE Trans Biomedical Engineering},
year = {2019},
volume = {66},
number = {7},
pages = {1949-1962},
url = {https://www.research-collection.ethz.ch/bitstream/handle/20.500.11850/310433/SoSwithreflector.pdf},
doi = {10.1109/TBME.2018.2881302}
}
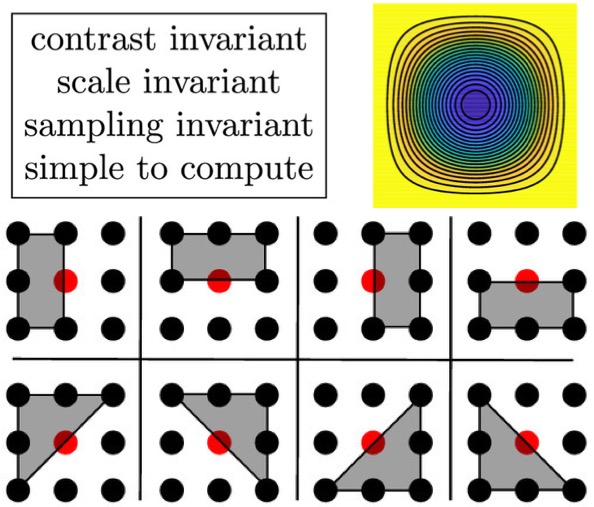 [2019] Yuanhao Gong and Orcun Goksel:
"Weighted Mean Curvature",
Signal Processing 164:329-339, Jun 2019.
[2019] Yuanhao Gong and Orcun Goksel:
"Weighted Mean Curvature",
Signal Processing 164:329-339, Jun 2019.
@article{Gong_weighted_19,
author = {Yuanhao Gong and Orcun Goksel},
title = {Weighted Mean Curvature},
journal = {Signal Processing},
year = {2019},
volume = {164},
pages = {329-339},
url = {https://arxiv.org/abs/1903.07189},
doi = {10.1016/j.sigpro.2019.06.020}
}
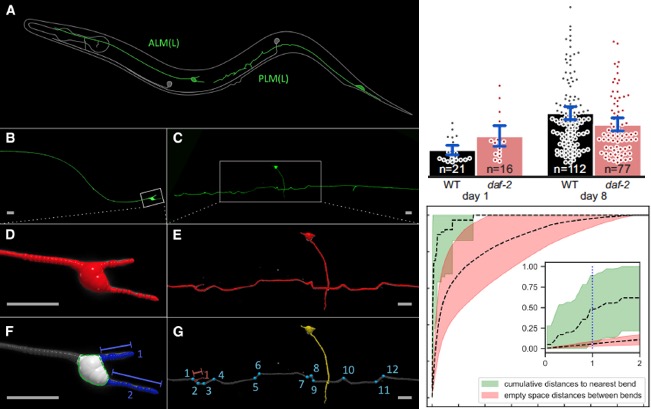 [2019] Max Hess, Alvaro Gomariz, Orcun Goksel, and Collin Ewald:
"In-vivo quantitative image analysis of age-related morphological changes of C. elegans neurons reveals a correlation between neurite bending and novel neurite outgrowths",
eNeuro 6(4), Jun 2019.
[2019] Max Hess, Alvaro Gomariz, Orcun Goksel, and Collin Ewald:
"In-vivo quantitative image analysis of age-related morphological changes of C. elegans neurons reveals a correlation between neurite bending and novel neurite outgrowths",
eNeuro 6(4), Jun 2019.
@article{Hess_in-vivo_19,
author = {Max Hess and Alvaro Gomariz and Orcun Goksel and Collin Ewald},
title = {In-vivo quantitative image analysis of age-related morphological changes of C. elegans neurons reveals a correlation between neurite bending and novel neurite outgrowths},
journal = {eNeuro},
year = {2019},
volume = {6},
number = {4},
doi = {10.1523/ENEURO.0014-19.2019}
}
 [2019] Lisa Ruby, Sergio J. Sanabria, Anika S. Obrist, Katharina Martini, Serafino Forte, Orcun Goksel, Thomas Frauenfelder, Rahel A. Kubik-Huch, and Marga B. Rominger:
"Breast Density Assessment in Young Women with Handheld Ultrasound Based on Speed of Sound: Influence of the Menstrual Cycle",
Medicine 98(25):e16123, Jun 2019.
[2019] Lisa Ruby, Sergio J. Sanabria, Anika S. Obrist, Katharina Martini, Serafino Forte, Orcun Goksel, Thomas Frauenfelder, Rahel A. Kubik-Huch, and Marga B. Rominger:
"Breast Density Assessment in Young Women with Handheld Ultrasound Based on Speed of Sound: Influence of the Menstrual Cycle",
Medicine 98(25):e16123, Jun 2019.
@article{Ruby_breast-density_19,
author = {Lisa Ruby and Sergio J Sanabria and Anika S Obrist and Katharina Martini and Serafino Forte and Orcun Goksel and Thomas Frauenfelder and Rahel A Kubik-Huch and Marga B Rominger},
title = {Breast Density Assessment in Young Women with Handheld Ultrasound Based on Speed of Sound: Influence of the Menstrual Cycle},
journal = {Medicine},
year = {2019},
volume = {98},
number = {25},
pages = {e16123},
doi = {10.1097/MD.0000000000016123}
}
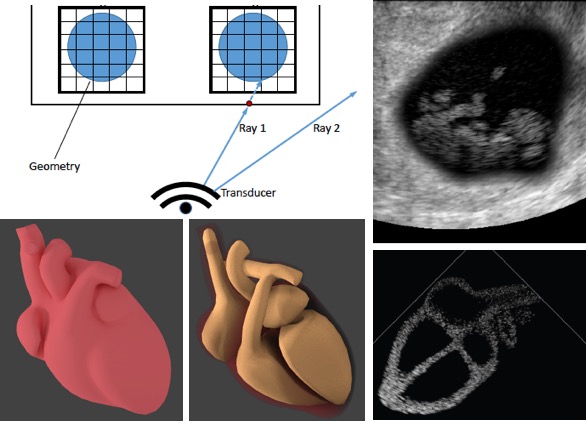 [2019] Rastislav Starkov, Christine Tanner, Michael Bajka, and Orcun Goksel:
"Ultrasound Simulation with Animated Anatomical Models and On-the-Fly Fusion with Real Images via Path Tracing",
Computers & Graphics 82:44-52, May 2019.
[2019] Rastislav Starkov, Christine Tanner, Michael Bajka, and Orcun Goksel:
"Ultrasound Simulation with Animated Anatomical Models and On-the-Fly Fusion with Real Images via Path Tracing",
Computers & Graphics 82:44-52, May 2019.
@article{Starkov_ultrasound_19,
author = {Rastislav Starkov and Christine Tanner and Michael Bajka and Orcun Goksel},
title = {Ultrasound Simulation with Animated Anatomical Models and On-the-Fly Fusion with Real Images via Path Tracing},
journal = {Computers & Graphics},
year = {2019},
volume = {82},
pages = {44-52},
url = {https://www.research-collection.ethz.ch/bitstream/handle/20.500.11850/345177/StarkovR_ComputGraph2019_UltrasoundAnimated.pdf},
doi = {10.1016/j.cag.2019.05.005}
}
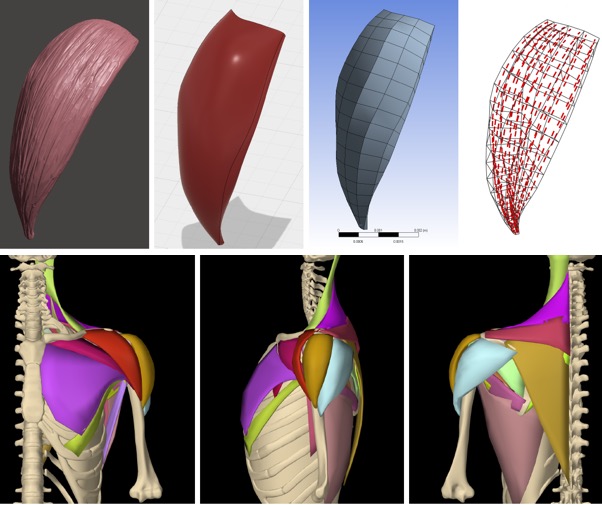 [2019] Fabien Pean, Christine Tanner, Christian Gerber, Philipp Fuernstahl, and Orcun Goksel:
"A comprehensive and volumetric musculoskeletal model for the dynamic simulation of the shoulder function",
Computer Methods in Biomechanics and Biomedical Engineering (CMBBE) 22(7):740-751, Apr 2019.
[2019] Fabien Pean, Christine Tanner, Christian Gerber, Philipp Fuernstahl, and Orcun Goksel:
"A comprehensive and volumetric musculoskeletal model for the dynamic simulation of the shoulder function",
Computer Methods in Biomechanics and Biomedical Engineering (CMBBE) 22(7):740-751, Apr 2019.
@article{Pean_comprehensive_19,
author = {Fabien Pean and Christine Tanner and Christian Gerber and Philipp Fuernstahl and Orcun Goksel},
title = {A comprehensive and volumetric musculoskeletal model for the dynamic simulation of the shoulder function},
journal = {Computer Methods in Biomechanics and Biomedical Engineering (CMBBE)},
year = {2019},
volume = {22},
number = {7},
pages = {740-751},
doi = {10.1080/10255842.2019.1588963}
}
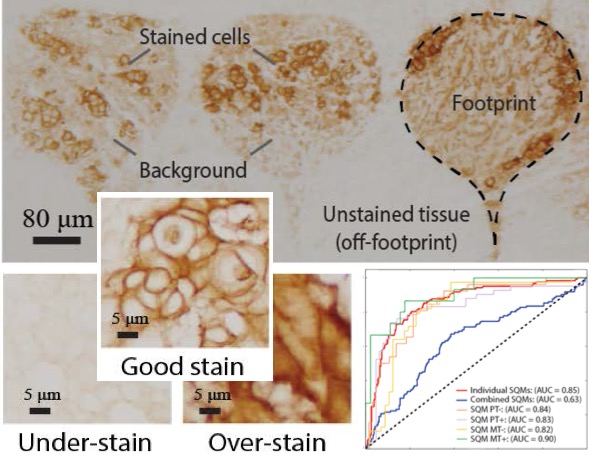 [2019] Nuri Murat Arar, Pushpak Pati, Aditya Kashyap, Anna Fomitcheva Khartchenko, Orcun Goksel, Govind V. Kaigala, and Maria Gabrani:
"High-Quality Immunohistochemical Stains through Computational Assay Parameter Optimization",
IEEE Trans Biomedical Engineering 66(10):2952-63, Feb 2019.
[2019] Nuri Murat Arar, Pushpak Pati, Aditya Kashyap, Anna Fomitcheva Khartchenko, Orcun Goksel, Govind V. Kaigala, and Maria Gabrani:
"High-Quality Immunohistochemical Stains through Computational Assay Parameter Optimization",
IEEE Trans Biomedical Engineering 66(10):2952-63, Feb 2019.
@article{Arar_high_19,
author = {Nuri Murat Arar and Pushpak Pati and Aditya Kashyap and Anna Fomitcheva Khartchenko and Orcun Goksel and Govind V. Kaigala and Maria Gabrani},
title = {High-Quality Immunohistochemical Stains through Computational Assay Parameter Optimization},
journal = {IEEE Trans Biomedical Engineering},
year = {2019},
volume = {66},
number = {10},
pages = {2952-63},
doi = {10.1109/TBME.2019.2899156}
}
 [2019] Torben Marhenke, Sergio J. Sanabria, Bhaskara Rao Chintada, Roman Furrer, Jürg Neuenschwander, and Orcun Goksel:
"Fast Spatial Characterization of Acoustic Fields Generated by Medical Array Transducers Based on Single-Plane Hydrophone Measurements",
Sensors 19(4):863, Feb 2019.
[2019] Torben Marhenke, Sergio J. Sanabria, Bhaskara Rao Chintada, Roman Furrer, Jürg Neuenschwander, and Orcun Goksel:
"Fast Spatial Characterization of Acoustic Fields Generated by Medical Array Transducers Based on Single-Plane Hydrophone Measurements",
Sensors 19(4):863, Feb 2019.
@article{Mahrenke_comprehensive_19,
author = {Torben Marhenke and Sergio J. Sanabria and Bhaskara Rao Chintada and Roman Furrer and J{\"u}rg Neuenschwander and Orcun Goksel},
title = {Fast Spatial Characterization of Acoustic Fields Generated by Medical Array Transducers Based on Single-Plane Hydrophone Measurements},
journal = {Sensors},
year = {2019},
volume = {19},
number = {4},
pages = {863},
doi = {10.3390/s19040863}
}
 [2019] Stefanie Ehrbar, Alexander Jöhl, Michael Kühni, Mirko Meboldt, Ece Ozkan Elsen, Christine Tanner, Orcun Goksel, Stephan Klöck, Jan Unkelbach, Matthias Guckenberger, and Stephanie Tanadini-Lang:
"ELPHA: Dynamically deformable liver phantom for real-time motion-adaptive radiotherapy treatments",
Medical Physics 46(2):839-850, Feb 2019.
[2019] Stefanie Ehrbar, Alexander Jöhl, Michael Kühni, Mirko Meboldt, Ece Ozkan Elsen, Christine Tanner, Orcun Goksel, Stephan Klöck, Jan Unkelbach, Matthias Guckenberger, and Stephanie Tanadini-Lang:
"ELPHA: Dynamically deformable liver phantom for real-time motion-adaptive radiotherapy treatments",
Medical Physics 46(2):839-850, Feb 2019.
@article{Ehrbar_elpha_19,
author = {Stefanie Ehrbar and Alexander J\"ohl and Michael K\"uhni and Mirko Meboldt and Ece Ozkan Elsen and Christine Tanner and Orcun Goksel and Stephan Kl\"ock and Jan Unkelbach and Matthias Guckenberger and Stephanie Tanadini-Lang},
title = {ELPHA: Dynamically deformable liver phantom for real-time motion-adaptive radiotherapy treatments},
journal = {Medical Physics},
year = {2019},
volume = {46},
number = {2},
pages = {839-850},
doi = {10.1002/mp.13359}
}
 [2019] Sergio J. Sanabria, Katharina Martini, Gregor Freystätter, Lisa Ruby, Orcun Goksel, Thomas Frauenfelder, and Marga B. Rominger:
"Speed of sound ultrasound: a pilot study on a novel technique to identify sarcopenia in seniors",
European Radiology 29(1):3-12, Jan 2019.
[2019] Sergio J. Sanabria, Katharina Martini, Gregor Freystätter, Lisa Ruby, Orcun Goksel, Thomas Frauenfelder, and Marga B. Rominger:
"Speed of sound ultrasound: a pilot study on a novel technique to identify sarcopenia in seniors",
European Radiology 29(1):3-12, Jan 2019.
@article{Sanabria_speed_19,
author = {Sergio J Sanabria and Katharina Martini and Gregor Freyst\"{a}tter and Lisa Ruby and Orcun Goksel and Thomas Frauenfelder and Marga B Rominger},
title = {Speed of sound ultrasound: a pilot study on a novel technique to identify sarcopenia in seniors},
journal = {European Radiology},
year = {2019},
volume = {29},
number = {1},
pages = {3-12},
doi = {10.1007/s00330-018-5742-2}
}
2018
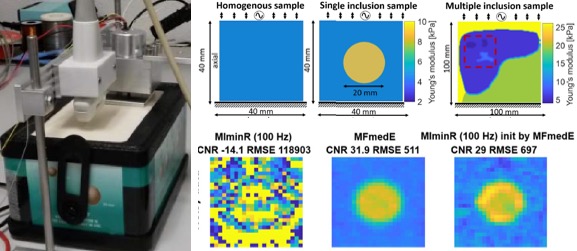 [2018] Corin F. Otesteanu, Sergio J. Sanabria, and Orcun Goksel:
"Robust Reconstruction of Elasticity Using Ultrasound Imaging and Multi-frequency Excitations",
IEEE Trans Medical Imaging 37(11):2502-2513, Nov 2018.
[2018] Corin F. Otesteanu, Sergio J. Sanabria, and Orcun Goksel:
"Robust Reconstruction of Elasticity Using Ultrasound Imaging and Multi-frequency Excitations",
IEEE Trans Medical Imaging 37(11):2502-2513, Nov 2018.
@article{Otesteanu_robust_18,
author = {Corin F Otesteanu and Sergio J Sanabria and Orcun Goksel},
title = {Robust Reconstruction of Elasticity Using Ultrasound Imaging and Multi-frequency Excitations},
journal = {IEEE Trans Medical Imaging},
year = {2018},
volume = {37},
number = {11},
pages = {2502-2513},
doi = {10.1109/TMI.2018.2837390}
}
 [2018] Sergio J. Sanabria, Ece Ozkan, Marga B. Rominger, and Orcun Goksel:
"Spatial Domain Reconstruction for Imaging Speed-of-Sound with Pulse-Echo Ultrasound: Simulation and In-Vivo Study",
Physics in Medicine and Biology 63:215015, Oct 2018.
[2018] Sergio J. Sanabria, Ece Ozkan, Marga B. Rominger, and Orcun Goksel:
"Spatial Domain Reconstruction for Imaging Speed-of-Sound with Pulse-Echo Ultrasound: Simulation and In-Vivo Study",
Physics in Medicine and Biology 63:215015, Oct 2018.
@article{Sanabria_spatial_18,
author = {Sergio J Sanabria and Ece Ozkan and Marga B Rominger and Orcun Goksel},
title = {Spatial Domain Reconstruction for Imaging Speed-of-Sound with Pulse-Echo Ultrasound: Simulation and In-Vivo Study},
journal = {Physics in Medicine and Biology},
year = {2018},
volume = {63},
pages = {215015},
doi = {10.1088/1361-6560/aae2fb}
}
 [2018] Valeria De Luca, Jyotirmoy Banerjee, Andre Hallack, Satoshi Kondo, Maxim Makhinya, Daniel Nouri, Lucas Royer, Amalia Cifor, Guillaume Dardenne, Orcun Goksel, Mark J. Gooding, Camiel Klink, Alexandre Krupa, Anthony Le Bras, Maud Marchal, Adriaan Moelker, Wiro J. Niessen, Bartlomiej W. Papiez, Alex Rothberg, Julia A. Schnabel, Theo van Walsum, Erwin Vast, Emma Harris, Muyinatu A. Lediju Bell, and Christine Tanner:
"Evaluation of 2D and 3D ultrasound tracking algorithms and impact on ultrasound-guided liver radiotherapy margins",
Medical Physics 45(11):4986-5003, Oct 2018.
[2018] Valeria De Luca, Jyotirmoy Banerjee, Andre Hallack, Satoshi Kondo, Maxim Makhinya, Daniel Nouri, Lucas Royer, Amalia Cifor, Guillaume Dardenne, Orcun Goksel, Mark J. Gooding, Camiel Klink, Alexandre Krupa, Anthony Le Bras, Maud Marchal, Adriaan Moelker, Wiro J. Niessen, Bartlomiej W. Papiez, Alex Rothberg, Julia A. Schnabel, Theo van Walsum, Erwin Vast, Emma Harris, Muyinatu A. Lediju Bell, and Christine Tanner:
"Evaluation of 2D and 3D ultrasound tracking algorithms and impact on ultrasound-guided liver radiotherapy margins",
Medical Physics 45(11):4986-5003, Oct 2018.
@article{DeLuca_evaluation_18,
author = {Valeria De Luca and Jyotirmoy Banerjee and Andre Hallack and Satoshi Kondo and Maxim Makhinya and Daniel Nouri and Lucas Royer and Amalia Cifor and Guillaume Dardenne and Orcun Goksel and Mark J. Gooding and Camiel Klink and Alexandre Krupa and Anthony Le Bras and Maud Marchal and Adriaan Moelker and Wiro J. Niessen and Bartlomiej W. Papiez and Alex Rothberg and Julia A. Schnabel and Theo van Walsum and Erwin Vast and Emma Harris and Muyinatu A. Lediju Bell and Christine Tanner},
title = {Evaluation of 2D and 3D ultrasound tracking algorithms and impact on ultrasound-guided liver radiotherapy margins},
journal = {Medical Physics},
year = {2018},
volume = {45},
number = {11},
pages = {4986-5003},
doi = {10.1002/mp.13152}
}
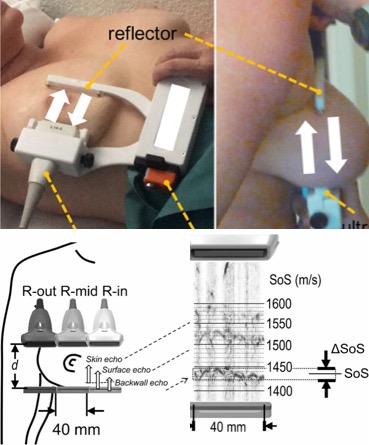 [2018] Sergio J. Sanabria, Orcun Goksel, Katharina Martini, Serafino Forte, Thomas Frauenfelder, Rahel A. Kubik-Huch, and Marga B. Rominger:
"Breast-Density Assessment with Handheld Ultrasound: A Novel Biomarker to Assess Breast Cancer Risk and to Tailor Screening?",
European Radiology 28(8):3165-3175, Aug 2018.
[2018] Sergio J. Sanabria, Orcun Goksel, Katharina Martini, Serafino Forte, Thomas Frauenfelder, Rahel A. Kubik-Huch, and Marga B. Rominger:
"Breast-Density Assessment with Handheld Ultrasound: A Novel Biomarker to Assess Breast Cancer Risk and to Tailor Screening?",
European Radiology 28(8):3165-3175, Aug 2018.
@article{Sanabria_breast-density_18,
author = {Sergio J Sanabria and Orcun Goksel and Katharina Martini and Serafino Forte and Thomas Frauenfelder and Rahel A Kubik-Huch and Marga B Rominger},
title = {Breast-Density Assessment with Handheld Ultrasound: A Novel Biomarker to Assess Breast Cancer Risk and to Tailor Screening?},
journal = {European Radiology},
year = {2018},
volume = {28},
number = {8},
pages = {3165-3175},
doi = {10.1007/s00330-017-5287-9}
}
 [2018] Golnoosh Samei, Orcun Goksel, Julio Lobo, Omid Mohareri, Peter Black, Robert Rohling, and Septimiu Salcudean:
"Real-time FEM-based Registration of 3D to 2.5D Transrectal Ultrasound Images",
IEEE Trans Medical Imaging 37(8):1877-86, Aug 2018.
[2018] Golnoosh Samei, Orcun Goksel, Julio Lobo, Omid Mohareri, Peter Black, Robert Rohling, and Septimiu Salcudean:
"Real-time FEM-based Registration of 3D to 2.5D Transrectal Ultrasound Images",
IEEE Trans Medical Imaging 37(8):1877-86, Aug 2018.
@article{Samei_real-time_18,
author = {Golnoosh Samei and Orcun Goksel and Julio Lobo and Omid Mohareri and Peter Black and Robert Rohling and Septimiu Salcudean},
title = {Real-time FEM-based Registration of 3D to 2.5D Transrectal Ultrasound Images},
journal = {IEEE Trans Medical Imaging},
year = {2018},
volume = {37},
number = {8},
pages = {1877-86},
doi = {10.1109/TMI.2018.2810778}
}
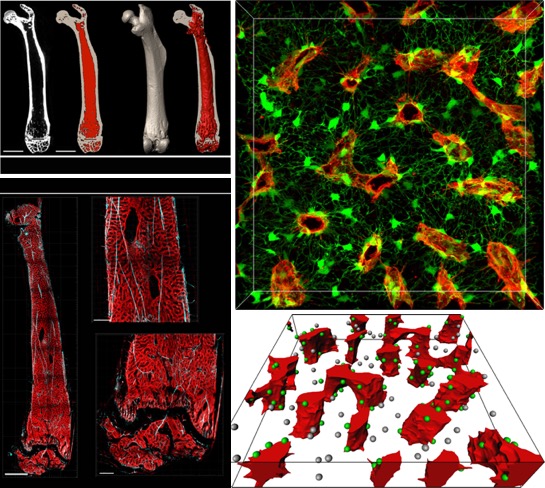 [2018] Alvaro Gomariz, Stephan Isringhausen, Patrick Helbling, Ute Suessbier, Anton Becker, Andreas Boss, Takashi Nagasawa, Grégory Paul, Orcun Goksel, Gábor Székely, Szymon Stoma, Simon F. N/orrelykke, Markus G. Manz, and César Nombela-Arrieta:
"Quantitative spatial analysis of hematopoiesis-regulating stromal cells in the bone marrow microenvironment by multiscale 3D microscopy",
Nature Communications 9(2532), Jun 2018.
[2018] Alvaro Gomariz, Stephan Isringhausen, Patrick Helbling, Ute Suessbier, Anton Becker, Andreas Boss, Takashi Nagasawa, Grégory Paul, Orcun Goksel, Gábor Székely, Szymon Stoma, Simon F. N/orrelykke, Markus G. Manz, and César Nombela-Arrieta:
"Quantitative spatial analysis of hematopoiesis-regulating stromal cells in the bone marrow microenvironment by multiscale 3D microscopy",
Nature Communications 9(2532), Jun 2018.
@article{Gomariz_quantitative_18,
author = {Alvaro Gomariz and Stephan Isringhausen and Patrick Helbling and Ute Suessbier and Anton Becker and Andreas Boss and Takashi Nagasawa and Gr\'egory Paul and Orcun Goksel and G\'abor Sz\'ekely and Szymon Stoma and Simon F. N\/orrelykke and Markus G. Manz and C\'esar Nombela-Arrieta},
title = {Quantitative spatial analysis of hematopoiesis-regulating stromal cells in the bone marrow microenvironment by multiscale 3D microscopy},
journal = {Nature Communications},
year = {2018},
volume = {9},
number = {2532},
doi = {10.1038/s41467-018-04770-z}
}
 [2018] Corin F. Otesteanu, Valeriy Vishnevskiy, and Orcun Goksel:
"FEM Based Elasticity Reconstruction Using Ultrasound For Imaging Tissue Ablation",
Int J Computer Assisted Radiology and Surgery 13(6):885-894, Jun 2018.
[2018] Corin F. Otesteanu, Valeriy Vishnevskiy, and Orcun Goksel:
"FEM Based Elasticity Reconstruction Using Ultrasound For Imaging Tissue Ablation",
Int J Computer Assisted Radiology and Surgery 13(6):885-894, Jun 2018.
@article{Otesteanu_fem_18,
author = {Corin F Otesteanu and Valeriy Vishnevskiy and Orcun Goksel},
title = {FEM Based Elasticity Reconstruction Using Ultrasound For Imaging Tissue Ablation},
journal = {Int J Computer Assisted Radiology and Surgery},
year = {2018},
volume = {13},
number = {6},
pages = {885-894},
doi = {10.1007/s11548-018-1714-x}
}
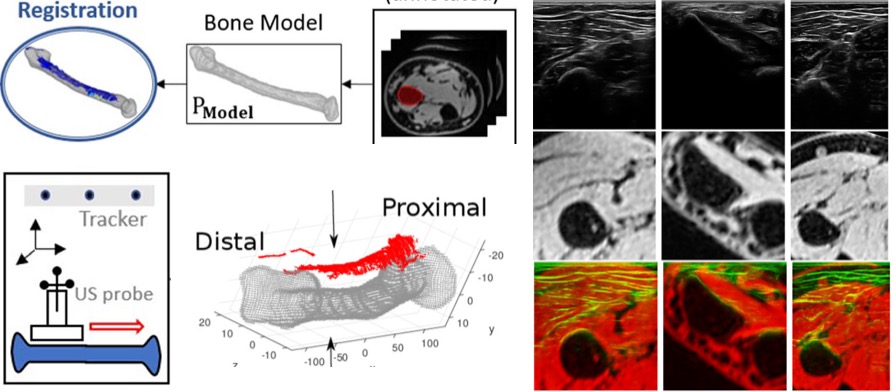 [2018] Matija Ciganovic, Firat Ozdemir, Fabien Pean, Philipp Fuernstahl, Christine Tanner, and Orcun Goksel:
"Registration of 3D Freehand Ultrasound to a Bone Model for Orthopaedic Procedures of the Forearm",
Int J Computer Assisted Radiology and Surgery 13(6):827-836, Jun 2018.
[2018] Matija Ciganovic, Firat Ozdemir, Fabien Pean, Philipp Fuernstahl, Christine Tanner, and Orcun Goksel:
"Registration of 3D Freehand Ultrasound to a Bone Model for Orthopaedic Procedures of the Forearm",
Int J Computer Assisted Radiology and Surgery 13(6):827-836, Jun 2018.
@article{Ciganovic_registration_18,
author = {Matija Ciganovic and Firat Ozdemir and Fabien Pean and Philipp Fuernstahl and Christine Tanner and Orcun Goksel},
title = {Registration of 3D Freehand Ultrasound to a Bone Model for Orthopaedic Procedures of the Forearm},
journal = {Int J Computer Assisted Radiology and Surgery},
year = {2018},
volume = {13},
number = {6},
pages = {827-836},
doi = {10.1007/s11548-018-1756-0}
}
 [2018] Oliver Mattausch and Orcun Goksel:
"Image-based Reconstruction of Tissue Scatterers using Beam Steering for Ultrasound Simulation",
IEEE Trans Medical Imaging 37(3):767-780, Mar 2018.
[2018] Oliver Mattausch and Orcun Goksel:
"Image-based Reconstruction of Tissue Scatterers using Beam Steering for Ultrasound Simulation",
IEEE Trans Medical Imaging 37(3):767-780, Mar 2018.
@article{Mattausch_image-based_18,
author = {Oliver Mattausch and Orcun Goksel},
title = {Image-based Reconstruction of Tissue Scatterers using Beam Steering for Ultrasound Simulation},
journal = {IEEE Trans Medical Imaging},
year = {2018},
volume = {37},
number = {3},
pages = {767-780},
doi = {10.1109/TMI.2017.2770118}
}
 [2018] Ece Ozkan, Valeriy Vishnevsky, and Orcun Goksel:
"Inverse Problem of Ultrasound Beamforming with Sparsity Constraints and Regularization",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 65(3):356-365, Mar 2018.
[2018] Ece Ozkan, Valeriy Vishnevsky, and Orcun Goksel:
"Inverse Problem of Ultrasound Beamforming with Sparsity Constraints and Regularization",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 65(3):356-365, Mar 2018.
@article{Ozkan_inverse_18,
author = {Ece Ozkan and Valeriy Vishnevsky and Orcun Goksel},
title = {Inverse Problem of Ultrasound Beamforming with Sparsity Constraints and Regularization},
journal = {IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control},
year = {2018},
volume = {65},
number = {3},
pages = {356-365},
doi = {10.1109/TUFFC.2017.2757880}
}
 [2018] Oliver Mattausch, Maxim Makhinya, and Orcun Goksel:
"Realistic Ultrasound Simulation of Complex Surface Models Using Interactive Monte-Carlo Path Tracing",
Computer Graphics Forum 37(1):202-213, Feb 2018.
[2018] Oliver Mattausch, Maxim Makhinya, and Orcun Goksel:
"Realistic Ultrasound Simulation of Complex Surface Models Using Interactive Monte-Carlo Path Tracing",
Computer Graphics Forum 37(1):202-213, Feb 2018.
@article{Mattausch_realistic_18,
author = {Oliver Mattausch and Maxim Makhinya and Orcun Goksel},
title = {Realistic Ultrasound Simulation of Complex Surface Models Using Interactive Monte-Carlo Path Tracing},
journal = {Computer Graphics Forum},
year = {2018},
volume = {37},
number = {1},
pages = {202-213},
doi = {10.1111/cgf.13260}
}
 [2018] Ece Ozkan and Orcun Goksel:
"Compliance Boundary Conditions for Patient-Specific Deformation Simulation Using the Finite Element Method",
Biomedical Physics & Engineering Express 4(2):025003, Jan 2018.
[2018] Ece Ozkan and Orcun Goksel:
"Compliance Boundary Conditions for Patient-Specific Deformation Simulation Using the Finite Element Method",
Biomedical Physics & Engineering Express 4(2):025003, Jan 2018.
@article{Ozkan_compliance_18,
author = {Ece Ozkan and Orcun Goksel},
title = {Compliance Boundary Conditions for Patient-Specific Deformation Simulation Using the Finite Element Method},
journal = {Biomedical Physics & Engineering Express},
year = {2018},
volume = {4},
number = {2},
pages = {025003},
doi = {10.1088/2057-1976/aa918d}
}
2017
 [2017] Elin L. Lundin, Martin Stauber, Panagiota Papageorgiou, Martin Ehrbar, Chafik Ghayord, Franz E. Weber, Christine Tanner, and Orcun Goksel:
"Automatic Registration of 2D Histological Sections to 3D microCT Volumes: Trabecular Bone",
Bone 105:173-183, Dec 2017.
[2017] Elin L. Lundin, Martin Stauber, Panagiota Papageorgiou, Martin Ehrbar, Chafik Ghayord, Franz E. Weber, Christine Tanner, and Orcun Goksel:
"Automatic Registration of 2D Histological Sections to 3D microCT Volumes: Trabecular Bone",
Bone 105:173-183, Dec 2017.
@article{Lundin_automatic_17,
author = {Elin L. Lundin and Martin Stauber and Panagiota Papageorgiou and Martin Ehrbar and Chafik Ghayord and Franz E. Weber and Christine Tanner and Orcun Goksel},
title = {Automatic Registration of 2D Histological Sections to 3D microCT Volumes: Trabecular Bone},
journal = {Bone},
year = {2017},
volume = {105},
pages = {173-183},
doi = {10.1016/j.bone.2017.08.021}
}
 [2017] Christine Tanner, Barbara Flach, Céline Eggenberger, Oliver Mattausch, Michael Bajka, and Orcun Goksel:
"Consistent Reconstruction of 4D Fetal Heart Ultrasound Images to Cope with Fetal Motion",
Int J Computer Assisted Radiology and Surgery 12(8):1307-17, Aug 2017.
[2017] Christine Tanner, Barbara Flach, Céline Eggenberger, Oliver Mattausch, Michael Bajka, and Orcun Goksel:
"Consistent Reconstruction of 4D Fetal Heart Ultrasound Images to Cope with Fetal Motion",
Int J Computer Assisted Radiology and Surgery 12(8):1307-17, Aug 2017.
@article{Tanner_consistent_17,
author = {Christine Tanner and Barbara Flach and C\'{e}line Eggenberger and Oliver Mattausch and Michael Bajka and Orcun Goksel},
title = {Consistent Reconstruction of 4D Fetal Heart Ultrasound Images to Cope with Fetal Motion},
journal = {Int J Computer Assisted Radiology and Surgery},
year = {2017},
volume = {12},
number = {8},
pages = {1307-17},
doi = {10.1007/s11548-017-1624-3}
}
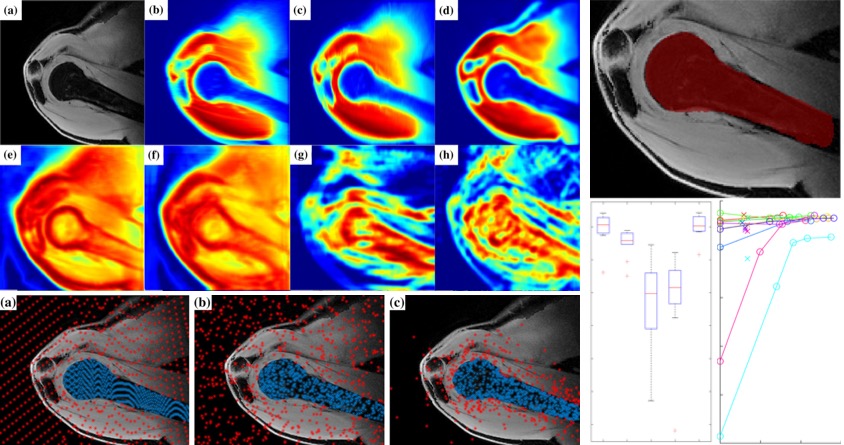 [2017] Firat Ozdemir, Neerav Karani, Philipp Fuernstahl, and Orcun Goksel:
"Interactive Segmentation in MRI for Orthopedic Surgery Planning: Bone Tissue",
Int J Comp. Assisted Radiol. Surgery 12(6):1031-9, Jun 2017.
[2017] Firat Ozdemir, Neerav Karani, Philipp Fuernstahl, and Orcun Goksel:
"Interactive Segmentation in MRI for Orthopedic Surgery Planning: Bone Tissue",
Int J Comp. Assisted Radiol. Surgery 12(6):1031-9, Jun 2017.
@article{Ozdemir_interactive_17,
author = {Firat Ozdemir and Neerav Karani and Philipp Fuernstahl and Orcun Goksel},
title = {Interactive Segmentation in MRI for Orthopedic Surgery Planning: Bone Tissue},
journal = {Int J Comp. Assisted Radiol. Surgery},
year = {2017},
volume = {12},
number = {6},
pages = {1031-9},
doi = {10.1007/s11548-017-1570-0}
}
 [2017] Ece Ozkan, Christine Tanner, Matej Kastelic, Oliver Mattausch, Maxim Makhinya, and Orcun Goksel:
"Robust Motion Tracking in Liver from 2D Ultrasound Images Using Supporters",
Int J Computer Assisted Radiology and Surgery 12(6):941-950, Jun 2017.
[2017] Ece Ozkan, Christine Tanner, Matej Kastelic, Oliver Mattausch, Maxim Makhinya, and Orcun Goksel:
"Robust Motion Tracking in Liver from 2D Ultrasound Images Using Supporters",
Int J Computer Assisted Radiology and Surgery 12(6):941-950, Jun 2017.
@article{Ozkan_robust_17,
author = {Ece Ozkan and Christine Tanner and Matej Kastelic and Oliver Mattausch and Maxim Makhinya and Orcun Goksel},
title = {Robust Motion Tracking in Liver from 2D Ultrasound Images Using Supporters},
journal = {Int J Computer Assisted Radiology and Surgery},
year = {2017},
volume = {12},
number = {6},
pages = {941-950},
doi = {10.1007/s11548-017-1559-8}
}
 [2017] Valeriy Vishnevskiy, Tobias Gass, Gabor Szekely, Christine Tanner, and Orcun Goksel:
"Isotropic Total Variation Regularization of Displacements in Parametric Image Registration",
IEEE Trans Medical Imaging 36(2):385-395, Feb 2017.
[2017] Valeriy Vishnevskiy, Tobias Gass, Gabor Szekely, Christine Tanner, and Orcun Goksel:
"Isotropic Total Variation Regularization of Displacements in Parametric Image Registration",
IEEE Trans Medical Imaging 36(2):385-395, Feb 2017.
@article{Vishnevskiy_isotropic_17,
author = {Valeriy Vishnevskiy and Tobias Gass and Gabor Szekely and Christine Tanner and Orcun Goksel},
title = {Isotropic Total Variation Regularization of Displacements in Parametric Image Registration},
journal = {IEEE Trans Medical Imaging},
year = {2017},
volume = {36},
number = {2},
pages = {385-395},
doi = {10.1109/tmi.2016.2610583}
}
2016 and Earlier
 [2016] Antje-Christin Knopf, Kristin Stützer, Christian Richter, Antoni Rucinsk, Joakim da Silva, Justin Phillips, Martijn Engelsman, Shinichi Shimizu, Rene Werner, Annika Jakobi, Orcun Goksel, Ye Zhang, Tuathan Oshea, Martin Fast, Rosalind Perrin, Christoph Bert, EriK Korevaar, and Jamie McClelland:
"Required transition from research to clinical application: report on the 4D treatment planning workshops 2014 and 2015",
Physica Medica: European J Medical Physics 32(7):874-82, Jul 2016.
[2016] Antje-Christin Knopf, Kristin Stützer, Christian Richter, Antoni Rucinsk, Joakim da Silva, Justin Phillips, Martijn Engelsman, Shinichi Shimizu, Rene Werner, Annika Jakobi, Orcun Goksel, Ye Zhang, Tuathan Oshea, Martin Fast, Rosalind Perrin, Christoph Bert, EriK Korevaar, and Jamie McClelland:
"Required transition from research to clinical application: report on the 4D treatment planning workshops 2014 and 2015",
Physica Medica: European J Medical Physics 32(7):874-82, Jul 2016.
@article{Knopf_required_16,
author = {Antje-Christin Knopf and Kristin St\"utzer and Christian Richter and Antoni Rucinsk and Joakim da Silva and Justin Phillips and Martijn Engelsman and Shinichi Shimizu and Rene Werner and Annika Jakobi and Orcun Goksel and Ye Zhang and Tuathan Oshea and Martin Fast and Rosalind Perrin and Christoph Bert and EriK Korevaar and Jamie McClelland},
title = {Required transition from research to clinical application: report on the 4D treatment planning workshops 2014 and 2015},
journal = {Physica Medica: European J Medical Physics},
year = {2016},
volume = {32},
number = {7},
pages = {874-82},
doi = {10.1016/j.ejmp.2016.05.064}
}
 [2016] Oscar Alfonso Jiménez-del-Toro, Henning Müller, Markus Krenn, Katharina Gruenberg, Abdel Aziz Taha, Marianne Winterstein, Ivan Eggel, Antonio Foncubierta-Rodríguez, Orcun Goksel, András Jakab, Georgios Kontokotsios, Georg Langs, Bjoern Menze, Tomàs Salas Fernandez, Roger Schaer, Anna Walley, Marc-Andr/e Weber, Yashin Dicente Cid, Tobias Gass, Mattias Heinrich, Fucang Jia, Fredrik Kahl, Razmig Kechichian, Dominic Mai, Assaf B. Spanier, Graham Vincent, Chunliang Wang, Daniel Wyeth, and Allan Hanbury:
"Cloud-based Evaluation of Anatomical Structure Segmentation and Landmark Detection Algorithms: VISCERAL Anatomy Benchmarks",
IEEE Trans Medical Imaging 35(11):2459-75, Nov 2016.
[2016] Oscar Alfonso Jiménez-del-Toro, Henning Müller, Markus Krenn, Katharina Gruenberg, Abdel Aziz Taha, Marianne Winterstein, Ivan Eggel, Antonio Foncubierta-Rodríguez, Orcun Goksel, András Jakab, Georgios Kontokotsios, Georg Langs, Bjoern Menze, Tomàs Salas Fernandez, Roger Schaer, Anna Walley, Marc-Andr/e Weber, Yashin Dicente Cid, Tobias Gass, Mattias Heinrich, Fucang Jia, Fredrik Kahl, Razmig Kechichian, Dominic Mai, Assaf B. Spanier, Graham Vincent, Chunliang Wang, Daniel Wyeth, and Allan Hanbury:
"Cloud-based Evaluation of Anatomical Structure Segmentation and Landmark Detection Algorithms: VISCERAL Anatomy Benchmarks",
IEEE Trans Medical Imaging 35(11):2459-75, Nov 2016.
@article{Jimenez_cloud-based_16,
author = {Oscar Alfonso {Jim\'enez-del-Toro} and Henning M\"uller and Markus Krenn and Katharina Gruenberg and Abdel Aziz Taha and Marianne Winterstein and Ivan Eggel and Antonio Foncubierta-Rodr\'iguez and Orcun Goksel and Andr\'as Jakab and Georgios Kontokotsios and Georg Langs and Bjoern Menze and Tom\`as Salas Fernandez and Roger Schaer and Anna Walley and Marc-Andr/e Weber and Yashin Dicente Cid and Tobias Gass and Mattias Heinrich and Fucang Jia and Fredrik Kahl and Razmig Kechichian and Dominic Mai and Assaf B. Spanier and Graham Vincent and Chunliang Wang and Daniel Wyeth and Allan Hanbury},
title = {Cloud-based Evaluation of Anatomical Structure Segmentation and Landmark Detection Algorithms: VISCERAL Anatomy Benchmarks},
journal = {IEEE Trans Medical Imaging},
year = {2016},
volume = {35},
number = {11},
pages = {2459-75},
doi = {10.1109/tmi.2016.2578680}
}
 [2016] Lazaros Vlachopoulos, Celestine Dünner, Tobias Gass, Matthias Graf, Orcun Goksel, Christian Gerber, Gábor Székely, and Philipp Fürnstahl:
"Computer algorithms for three-dimensional measurement of humeral anatomy: analysis of 140 paired humeri",
J Shoulder and Elbow Surgery 25(2):e38-e48, Feb 2016.
[2016] Lazaros Vlachopoulos, Celestine Dünner, Tobias Gass, Matthias Graf, Orcun Goksel, Christian Gerber, Gábor Székely, and Philipp Fürnstahl:
"Computer algorithms for three-dimensional measurement of humeral anatomy: analysis of 140 paired humeri",
J Shoulder and Elbow Surgery 25(2):e38-e48, Feb 2016.
@article{Vlachopoulos_computer_16,
author = {Lazaros Vlachopoulos and Celestine D\"unner and Tobias Gass and Matthias Graf and Orcun Goksel and Christian Gerber and G\'abor Sz\'ekely and Philipp F\"urnstahl},
title = {Computer algorithms for three-dimensional measurement of humeral anatomy: analysis of 140 paired humeri},
journal = {J Shoulder and Elbow Surgery},
year = {2016},
volume = {25},
number = {2},
pages = {e38-e48},
doi = {10.1016/j.jse.2015.07.027}
}
 [2016] Alessandro Crimi, Maxim Makhinya, Ulrich Baumann, Christoph Thalhammer, Gabor Szekely, and Orcun Goksel:
"Automatic Measurement of Venous Pressure Using B-Mode Ultrasound",
IEEE Trans Biomedical Engineering 63(2):288-299, Feb 2016.
[2016] Alessandro Crimi, Maxim Makhinya, Ulrich Baumann, Christoph Thalhammer, Gabor Szekely, and Orcun Goksel:
"Automatic Measurement of Venous Pressure Using B-Mode Ultrasound",
IEEE Trans Biomedical Engineering 63(2):288-299, Feb 2016.
* Having been selected as the best project of the year, this work received 2014 CTI MedTech Award at the funding agency's annual event.
@article{Crimi_automatic_16,
author = {Alessandro Crimi and Maxim Makhinya and Ulrich Baumann and Christoph Thalhammer and Gabor Szekely and Orcun Goksel},
title = {Automatic Measurement of Venous Pressure Using B-Mode Ultrasound},
journal = {IEEE Trans Biomedical Engineering},
year = {2016},
volume = {63},
number = {2},
pages = {288-299},
doi = {10.1109/TBME.2015.2455953}
}
 [2015] Siavash Khallaghi, C. Antonio Sánchez, Joy Sun, Farhad Imani, Amir Khojaste Galesh Khale, Orcun Goksel, Abtin Rasoulian, Cesare Romagnoli, Hamidreza Abdi, Silvia Chang, Parvin Mousavi, Aaron Fenster, Aaron Ward, Sidney Fels, and Purang Abolmaesumi:
"Biomechanically Constrained Surface Registration: Application to MR-TRUS Fusion for Prostate Interventions",
IEEE Trans Med Imag 34(11):2404-14, Nov 2015.
[2015] Siavash Khallaghi, C. Antonio Sánchez, Joy Sun, Farhad Imani, Amir Khojaste Galesh Khale, Orcun Goksel, Abtin Rasoulian, Cesare Romagnoli, Hamidreza Abdi, Silvia Chang, Parvin Mousavi, Aaron Fenster, Aaron Ward, Sidney Fels, and Purang Abolmaesumi:
"Biomechanically Constrained Surface Registration: Application to MR-TRUS Fusion for Prostate Interventions",
IEEE Trans Med Imag 34(11):2404-14, Nov 2015.
@article{Khallaghi_biomechanically_15,
author = {Siavash Khallaghi and C. Antonio S\'{a}nchez and Joy Sun and Farhad Imani and Amir Khojaste Galesh Khale and Orcun Goksel and Abtin Rasoulian and Cesare Romagnoli and Hamidreza Abdi and Silvia Chang and Parvin Mousavi and Aaron Fenster and Aaron Ward and Sidney Fels and Purang Abolmaesumi},
title = {Biomechanically Constrained Surface Registration: Application to MR-TRUS Fusion for Prostate Interventions},
journal = {IEEE Trans Med Imag},
year = {2015},
volume = {34},
number = {11},
pages = {2404-14},
doi = {10.1109/TMI.2015.2440253}
}
 [2015] Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Consistency-Based Rectification of Non-Rigid Registrations",
SPIE J Medical Imaging 2(1):014005, May 2015.
[2015] Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Consistency-Based Rectification of Non-Rigid Registrations",
SPIE J Medical Imaging 2(1):014005, May 2015.
@article{Gass_consistency-based_15,
author = {Tobias Gass and Gabor Szekely and Orcun Goksel},
title = {Consistency-Based Rectification of Non-Rigid Registrations},
journal = {SPIE J Medical Imaging},
year = {2015},
volume = {2},
number = {1},
pages = {014005},
doi = {10.1117/1.JMI.2.1.014005}
}
 [2015] Peter Baki, Sergio J. Sanabria, Gabor Kosa, Gabor Szekely, and Orcun Goksel:
"Thermal Expansion Imaging for Monitoring Lesion Depth using M-Mode Ultrasound during Cardiac RF Ablation: In-vitro Study",
Int J Computer Assisted Radiology and Surgery 10(6):681-693, Jun 2015.
[2015] Peter Baki, Sergio J. Sanabria, Gabor Kosa, Gabor Szekely, and Orcun Goksel:
"Thermal Expansion Imaging for Monitoring Lesion Depth using M-Mode Ultrasound during Cardiac RF Ablation: In-vitro Study",
Int J Computer Assisted Radiology and Surgery 10(6):681-693, Jun 2015.
* Received 3rd place (of 100 papers) in IPCAI/PHILIPS Best Paper Award at Information Processing in Computer-Assisted Interventions Conference, Barcelona, Spain, June 2015
@article{Baki_thermal_15,
author = {Peter Baki and Sergio J Sanabria and Gabor Kosa and Gabor Szekely and Orcun Goksel},
title = {Thermal Expansion Imaging for Monitoring Lesion Depth using M-Mode Ultrasound during Cardiac RF Ablation: In-vitro Study},
journal = {Int J Computer Assisted Radiology and Surgery},
year = {2015},
volume = {10},
number = {6},
pages = {681-693},
doi = {10.1007/s11548-015-1203-4}
}
 [2014] Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Simultaneous Segmentation and Multi-Resolution Nonrigid Atlas Registration",
IEEE Trans Image Processing 23(7):2931-43, July 2014.
[2014] Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Simultaneous Segmentation and Multi-Resolution Nonrigid Atlas Registration",
IEEE Trans Image Processing 23(7):2931-43, July 2014.
@article{Gass_simultaneous_14,
author = {Tobias Gass and Gabor Szekely and Orcun Goksel},
title = {Simultaneous Segmentation and Multi-Resolution Nonrigid Atlas Registration},
journal = {IEEE Trans Image Processing},
year = {2014},
volume = {23},
number = {7},
pages = {2931-43},
doi = {10.1109/TIP.2014.2322447}
}
 [2013] Orcun Goksel, Kirill Sapchuk, William James Morris, and Septimiu E. Salcudean:
"Prostate Brachytherapy Training with Simulated Ultrasound and Fluoroscopy Images",
IEEE Trans Biomedical Engineering 60(4):1002-12, Apr 2013.
[2013] Orcun Goksel, Kirill Sapchuk, William James Morris, and Septimiu E. Salcudean:
"Prostate Brachytherapy Training with Simulated Ultrasound and Fluoroscopy Images",
IEEE Trans Biomedical Engineering 60(4):1002-12, Apr 2013.
@article{Goksel_prostate_13,
author = {Orcun Goksel and Kirill Sapchuk and William James Morris and Septimiu E. Salcudean},
title = {Prostate Brachytherapy Training with Simulated Ultrasound and Fluoroscopy Images},
journal = {IEEE Trans Biomedical Engineering},
year = {2013},
volume = {60},
number = {4},
pages = {1002-12},
doi = {10.1109/TBME.2012.2222642}
}
 [2013] Orcun Goksel, Hani Eskandari, and Septimiu E. Salcudean:
"Mesh Adaptation for Improving Elasticity Reconstruction using the FEM Inverse Problem",
IEEE Trans Medical Imaging 32(2):408-418, Feb 2013.
[2013] Orcun Goksel, Hani Eskandari, and Septimiu E. Salcudean:
"Mesh Adaptation for Improving Elasticity Reconstruction using the FEM Inverse Problem",
IEEE Trans Medical Imaging 32(2):408-418, Feb 2013.
@article{Goksel_mesh_13,
author = {Orcun Goksel and Hani Eskandari and Septimiu E. Salcudean},
title = {Mesh Adaptation for Improving Elasticity Reconstruction using the FEM Inverse Problem},
journal = {IEEE Trans Medical Imaging},
year = {2013},
volume = {32},
number = {2},
pages = {408-418},
doi = {10.1109/TMI.2012.2228664}
}
 [2012] Marcel Lüthi, Remi Blanc, Thomas Albrecht, Tobias Gass, Orcun Goksel, Philippe Büchler, Michael Kistler, Habib Bousleiman, Mauricio Reyes, Philippe Cattin, and Thomas Vetter:
"Statismo - A framework for PCA based statistical models",
Insight Journal, Jul 2012.
[2012] Marcel Lüthi, Remi Blanc, Thomas Albrecht, Tobias Gass, Orcun Goksel, Philippe Büchler, Michael Kistler, Habib Bousleiman, Mauricio Reyes, Philippe Cattin, and Thomas Vetter:
"Statismo - A framework for PCA based statistical models",
Insight Journal, Jul 2012.
@article{Luethi_statismo_12,
author = {Marcel L\"uthi and Remi Blanc and Thomas Albrecht and Tobias Gass and Orcun Goksel and Philippe B\"uchler and Michael Kistler and Habib Bousleiman and Mauricio Reyes and Philippe Cattin and Thomas Vetter},
title = {Statismo - A framework for PCA based statistical models},
journal = {Insight Journal},
year = {2012},
url = {http://hdl.handle.net/10380/3371}
}
 [2012] Hani Eskandari, Orcun Goksel, Septimiu E. Salcudean, and Robert Rohling:
"Dilatation parameterization for two dimensional modeling of nearly incompressible isotropic materials",
Physics in Medicine and Biology 57(12):4055-73, Jun 2012.
[2012] Hani Eskandari, Orcun Goksel, Septimiu E. Salcudean, and Robert Rohling:
"Dilatation parameterization for two dimensional modeling of nearly incompressible isotropic materials",
Physics in Medicine and Biology 57(12):4055-73, Jun 2012.
@article{Eskandari_dilatation_12,
author = {Hani Eskandari and Orcun Goksel and Septimiu E. Salcudean and Robert Rohling},
title = {Dilatation parameterization for two dimensional modeling of nearly incompressible isotropic materials},
journal = {Physics in Medicine and Biology},
year = {2012},
volume = {57},
number = {12},
pages = {4055-73},
doi = {10.1088/0031-9155/57/12/4055}
}
 [2012] Jeffrey M. Abeysekera, Reza Zahiri-Azar, Orcun Goksel, Robert Rohling, and Septimiu E. Salcudean:
"Analysis of 2-D Motion Tracking in Ultrasound with Dual Transducers",
Ultrasonics 52(1):156-168, Jan 2012.
[2012] Jeffrey M. Abeysekera, Reza Zahiri-Azar, Orcun Goksel, Robert Rohling, and Septimiu E. Salcudean:
"Analysis of 2-D Motion Tracking in Ultrasound with Dual Transducers",
Ultrasonics 52(1):156-168, Jan 2012.
@article{Abeysekera_analysis_12,
author = {Jeffrey M. Abeysekera and Reza Zahiri-Azar and Orcun Goksel and Robert Rohling and Septimiu E. Salcudean},
title = {Analysis of 2-D Motion Tracking in Ultrasound with Dual Transducers},
journal = {Ultrasonics},
year = {2012},
volume = {52},
number = {1},
pages = {156-168},
doi = {10.1016/j.ultras.2011.07.011}
}
 [2011] Reza Zahiri Azar, Orcun Goksel, and Septimiu E. Salcudean:
"Comparison Between 2-D Cross Correlation With 2-D Sub-Sampling and 2-D Tracking Using Beam Steering",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 58(8):1534-7, Aug 2011.
[2011] Reza Zahiri Azar, Orcun Goksel, and Septimiu E. Salcudean:
"Comparison Between 2-D Cross Correlation With 2-D Sub-Sampling and 2-D Tracking Using Beam Steering",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 58(8):1534-7, Aug 2011.
@article{Azar_comparison_11,
author = {Reza Zahiri Azar and Orcun Goksel and Septimiu E. Salcudean},
title = {Comparison Between 2-D Cross Correlation With 2-D Sub-Sampling and 2-D Tracking Using Beam Steering},
journal = {IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control},
year = {2011},
volume = {58},
number = {8},
pages = {1534-7},
doi = {10.1109/TUFFC.2011.1978}
}
 [2011] Hani Eskandari, Orcun Goksel, Septimiu E. Salcudean, and Robert Rohling:
"Bandpass Sampling of High Frequency Tissue Motion",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 58(7):1332-43, Jul 2011.
[2011] Hani Eskandari, Orcun Goksel, Septimiu E. Salcudean, and Robert Rohling:
"Bandpass Sampling of High Frequency Tissue Motion",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 58(7):1332-43, Jul 2011.
@article{Eskandari_bandpass_11,
author = {Hani Eskandari and Orcun Goksel and Septimiu E. Salcudean and Robert Rohling},
title = {Bandpass Sampling of High Frequency Tissue Motion},
journal = {IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control},
year = {2011},
volume = {58},
number = {7},
pages = {1332-43},
doi = {10.1109/TUFFC.2011.1953}
}
 [2011] Orcun Goksel, Kirill Sapchuk, and Septimiu E. Salcudean:
"Haptic Simulator for Prostate Brachytherapy with Simulated Needle and Probe Interaction",
IEEE Trans Haptics 4(3):188-198, May 2011.
[2011] Orcun Goksel, Kirill Sapchuk, and Septimiu E. Salcudean:
"Haptic Simulator for Prostate Brachytherapy with Simulated Needle and Probe Interaction",
IEEE Trans Haptics 4(3):188-198, May 2011.
* Featured on the issue cover page
@article{Goksel_haptic_11,
author = {Orcun Goksel and Kirill Sapchuk and Septimiu E. Salcudean},
title = {Haptic Simulator for Prostate Brachytherapy with Simulated Needle and Probe Interaction},
journal = {IEEE Trans Haptics},
year = {2011},
volume = {4},
number = {3},
pages = {188-198},
doi = {10.1109/TOH.2011.34}
}
 [2011] Orcun Goksel and Septimiu E. Salcudean:
"Image-Based Variational Meshing",
IEEE Trans Medical Imaging 30(1):11-21, Jan 2011.
[2011] Orcun Goksel and Septimiu E. Salcudean:
"Image-Based Variational Meshing",
IEEE Trans Medical Imaging 30(1):11-21, Jan 2011.
@article{Goksel_image-based_11,
author = {Orcun Goksel and Septimiu E. Salcudean},
title = {Image-Based Variational Meshing},
journal = {IEEE Trans Medical Imaging},
year = {2011},
volume = {30},
number = {1},
pages = {11-21},
doi = {10.1109/TMI.2010.2055884}
}
 [2010] Reza Zahiri-Azar, Orcun Goksel, and Septimiu E. Salcudean:
"Sub-sample Displacement Estimation from Digitized Ultrasound RF Signals Using Multi-Dimensional Polynomial Fitting of the Cross-correlation Function",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 57(11):2403-20, Nov 2010.
[2010] Reza Zahiri-Azar, Orcun Goksel, and Septimiu E. Salcudean:
"Sub-sample Displacement Estimation from Digitized Ultrasound RF Signals Using Multi-Dimensional Polynomial Fitting of the Cross-correlation Function",
IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control 57(11):2403-20, Nov 2010.
@article{Zahiri-Azar_sub-sample_10,
author = {Reza Zahiri-Azar and Orcun Goksel and Septimiu E. Salcudean},
title = {Sub-sample Displacement Estimation from Digitized Ultrasound RF Signals Using Multi-Dimensional Polynomial Fitting of the Cross-correlation Function},
journal = {IEEE Trans Ultrasonics, Ferroelectrics, and Frequency Control},
year = {2010},
volume = {57},
number = {11},
pages = {2403-20},
doi = {10.1109/TUFFC.2010.1708}
}
 [2009] Orcun Goksel and Septimiu E. Salcudean:
"B-Mode Ultrasound Image Simulation in Deformable 3-D Medium",
IEEE Trans Medical Imaging 28(11):1657-69, Nov 2009.
[2009] Orcun Goksel and Septimiu E. Salcudean:
"B-Mode Ultrasound Image Simulation in Deformable 3-D Medium",
IEEE Trans Medical Imaging 28(11):1657-69, Nov 2009.
@article{Goksel_b-mode_09,
author = {Orcun Goksel and Septimiu E. Salcudean},
title = {B-Mode Ultrasound Image Simulation in Deformable 3-D Medium},
journal = {IEEE Trans Medical Imaging},
year = {2009},
volume = {28},
number = {11},
pages = {1657-69},
doi = {10.1109/TMI.2009.2016561}
}
 [2009] Orcun Goksel, Ehsan Dehghan, and Septimiu E. Salcudean:
"Modeling and Simulation of Flexible Needles",
Medical Engineering and Physics 31(9):1069-78, Nov 2009.
[2009] Orcun Goksel, Ehsan Dehghan, and Septimiu E. Salcudean:
"Modeling and Simulation of Flexible Needles",
Medical Engineering and Physics 31(9):1069-78, Nov 2009.
@article{Goksel_modeling_09,
author = {Orcun Goksel and Ehsan Dehghan and Septimiu E. Salcudean},
title = {Modeling and Simulation of Flexible Needles},
journal = {Medical Engineering and Physics},
year = {2009},
volume = {31},
number = {9},
pages = {1069-78},
doi = {10.1016/j.medengphy.2009.07.007}
}
 [2006] Orcun Goksel, Septimiu E. Salcudean, and Simon P. DiMaio:
"3D Simulation of Needle-Tissue Interaction with Application to Prostate Brachytherapy",
Computer Aided Surgery 11(6):279-288, Nov 2006.
[2006] Orcun Goksel, Septimiu E. Salcudean, and Simon P. DiMaio:
"3D Simulation of Needle-Tissue Interaction with Application to Prostate Brachytherapy",
Computer Aided Surgery 11(6):279-288, Nov 2006.
@article{Goksel_3d_06,
author = {Orcun Goksel and Septimiu E. Salcudean and Simon P. DiMaio},
title = {3D Simulation of Needle-Tissue Interaction with Application to Prostate Brachytherapy},
journal = {Computer Aided Surgery},
year = {2006},
volume = {11},
number = {6},
pages = {279-288},
doi = {10.3109/10929080601089997}
}
 [2005] Danny G. French, James Morris, Mira Keyes, Orcun Goksel, and Septimiu E. Salcudean:
"Computing Intraoperative Dosimetry for Prostate Brachytherapy Using TRUS and Fluoroscopy",
Academic Radiology 12(10):1262-72, Oct 2005.
[2005] Danny G. French, James Morris, Mira Keyes, Orcun Goksel, and Septimiu E. Salcudean:
"Computing Intraoperative Dosimetry for Prostate Brachytherapy Using TRUS and Fluoroscopy",
Academic Radiology 12(10):1262-72, Oct 2005.
@article{French_computing_05,
author = {Danny G. French and James Morris and Mira Keyes and Orcun Goksel and Septimiu E. Salcudean},
title = {Computing Intraoperative Dosimetry for Prostate Brachytherapy Using TRUS and Fluoroscopy},
journal = {Academic Radiology},
year = {2005},
volume = {12},
number = {10},
pages = {1262-72},
doi = {10.1016/j.acra.2005.05.026}
}
Book Chapters:
[2022] Pushpak Pati, Guillaume Jaume, Antonio Foncubierta-Rodriguez, Florinda Feroce, Giosue Scognamiglio, Anna Maria Anniciello, Nadia Brancati, Maria Frucci, Daniel Riccio, Jean-Philippe Thiran, Orcun Goksel, and Maria Gabrani: "Graph Representation Learning and Explainability in Breast Cancer Pathology: Bridging the Gap between AI and Pathology Practice", In Artificial Intelligence Applications In Human Pathology, pp. 243-285, World Scientific Publishing Europe, 2022.
@incollection{Pati_graph_22,
author = {Pushpak Pati and Guillaume Jaume and Antonio Foncubierta-Rodriguez and Florinda Feroce and Giosue Scognamiglio and Anna Maria Anniciello and Nadia Brancati and Maria Frucci and Daniel Riccio and Jean-Philippe Thiran and Orcun Goksel and Maria Gabrani},
title = {Graph Representation Learning and Explainability in Breast Cancer Pathology: Bridging the Gap between AI and Pathology Practice},
booktitle = {Artificial Intelligence Applications In Human Pathology},
publisher = {World Scientific Publishing Europe},
year = {2022},
pages = {243-285},
doi = {10.1142/9781800611399_0010}
}
[2017] Orcun Goksel and Antonio Foncubierta-Rodr\iguez: "VISCERAL Anatomy Benchmarks for Organ Segmentation and Landmark Localization: Tasks and Results", In Cloud-Based Benchmarking of Medical Image Analysis, pp. 107-125, Springer, 2017.
@incollection{Goksel_visceral_17,
author = {Goksel, Orcun and Foncubierta-Rodr{\'\i}guez, Antonio},
title = {VISCERAL Anatomy Benchmarks for Organ Segmentation and Landmark Localization: Tasks and Results},
booktitle = {Cloud-Based Benchmarking of Medical Image Analysis},
publisher = {Springer},
year = {2017},
pages = {107--125},
doi = {10.1007/978-3-319-49644-3_7}
}
[2016] Philipp Fürnstahl, Andreas Schweizer, Matthias Graf, Lazaros Vlachopoulos, Sandro Fucentese, Stephan Wirth, Ladislav Nagy, Gabor Szekely, and Orcun Goksel: "Surgical Treatment of Long-bone Deformities: 3D Preoperative Planning and Patient-specific Instrumentation", In Computational Radiology for Orthopaedic Interventions, ed. Guoyan Zheng and Shuo Li, pp. 123-149, Springer, 2016.
@incollection{Fuernstahl_surgical_16,
author = {Philipp F\"urnstahl and Andreas Schweizer and Matthias Graf and Lazaros Vlachopoulos and Sandro Fucentese and Stephan Wirth and Ladislav Nagy and Gabor Szekely and Orcun Goksel},
title = {Surgical Treatment of Long-bone Deformities: 3D Preoperative Planning and Patient-specific Instrumentation},
booktitle = {Computational Radiology for Orthopaedic Interventions},
editor = {Guoyan Zheng and Shuo Li},
publisher = {Springer},
year = {2016},
pages = {123-149},
url = {http://www.springer.com/us/book/9783319234816},
doi = {10.1007/978-3-319-23482-3_7}
}
[2014] Orçun Göksel and Gabor Székely: "Computational Support for Intraoperative Imaging and IGT", In Intraoperative Imaging and Image-Guided Therapy, ed. Ferenc A. Jolesz, pp. 63-77, Springer, New York, Jan 2014.
@incollection{Goksel_computational_14,
author = {Or\c{c}un G\"oksel and Gabor Sz\'ekely},
title = {Computational Support for Intraoperative Imaging and IGT},
booktitle = {Intraoperative Imaging and Image-Guided Therapy},
editor = {Ferenc A. Jolesz},
publisher = {Springer},
year = {2014},
pages = {63-77},
doi = {10.1007/978-1-4614-7657-3_4}
}
[2012] Septimiu E. Salcudean, Ramin S. Sahebjavaher, Orcun Goksel, Ali Baghani, Sara S. Mahdavi, Guy Nir, Ralph Sinkus, and Mehdi Moradi: "Biomechanical Modeling of the Prostate for Procedure Guidance and Simulation", In Soft Tissue Biomechanical Modeling for Computer Assisted Surgery, ed. Yohan Payan, pp. 169-198, v 11, Springer, Heidelberg, 2012.
@incollection{Salcudean_biomechanical_12,
author = {Septimiu E. Salcudean and Ramin S. Sahebjavaher and Orcun Goksel and Ali Baghani and Sara S. Mahdavi and Guy Nir and Ralph Sinkus and Mehdi Moradi},
title = {Biomechanical Modeling of the Prostate for Procedure Guidance and Simulation},
booktitle = {Soft Tissue Biomechanical Modeling for Computer Assisted Surgery},
editor = {Yohan Payan},
publisher = {Springer},
year = {2012},
volume = {11},
pages = {169-198},
doi = {10.1007/978-3-642-29014-5}
}
[2015] Orcun Goksel, Antonio Foncubierta-Rodríguez, Oscar Alfonso Jiménez del Toro, Henning Müller, Georg Langs, Marc-André Weber, Bjoern Menze, Ivan Eggel, Katharina Gruenberg, Marianne Winterstein, Markus Holzer, Markus Krenn, Georgios Kontokotsios, Sokratis Metallidis, Roger Schaer, Abdel Aziz Taha, András Jakab, Tomàs Salas Fernandez, and Allan Hanbury: "Overview of the VISCERAL Challenge at ISBI 2015", In Proceedings of VISCERAL Challenge at ISBI, ed. Orcun Goksel et al., pp. 6-11 (1390), CEUR-WS, Apr 2015.
@incollection{Goksel_overview_15,
author = {Orcun Goksel and Antonio Foncubierta--Rodr\'iguez and Oscar Alfonso Jim\'enez del Toro and Henning M\"uller and Georg Langs and Marc-Andr\'e Weber and Bjoern Menze and Ivan Eggel and Katharina Gruenberg and Marianne Winterstein and Markus Holzer and Markus Krenn and Georgios Kontokotsios and Sokratis Metallidis and Roger Schaer and Abdel Aziz Taha and Andr\'as Jakab and Tom\`as Salas Fernandez and Allan Hanbury},
title = {Overview of the VISCERAL Challenge at ISBI 2015},
booktitle = {Proceedings of VISCERAL Challenge at ISBI},
editor = {Orcun Goksel et al.},
publisher = {CEUR-WS},
year = {2015},
number = {1390},
pages = {6-11},
url = {http://ceur-ws.org/Vol-1390/visceralISBI15-0.pdf}
}
[2014] Oscar Alfonso Jiménez del Toro, Orcun Goksel, Bjoern Menze, Henning Müller, Georg Langs, Marc-André Weber, Ivan Eggel, Katharina Gruenberg, Markus Holzer, András Jakab, Georgios Kotsios-Kontokotsios, Markus Krenn, Tomàs Salas Fernandez, Roger Schaer, Abdel Aziz Taha, Marianne Winterstein, and Allan Hanbury: "VISCERAL - VISual Concept Extraction challenge in RAdioLogy : ISBI 2014 Challenge Organization", In VISCERAL Challenge at ISBI, ed. Orcun Goksel, pp. 6-15 (1194), CEUR-WS, May 2014.
@incollection{Jimenez_visceral_14,
author = {Jim\'enez del Toro, Oscar Alfonso and Orcun Goksel and Bjoern Menze and Henning M\"uller and Georg Langs and Marc-Andr\'e Weber and Ivan Eggel and Katharina Gruenberg and Markus Holzer and Andr\'as Jakab and Georgios Kotsios-Kontokotsios and Markus Krenn and Tom\`as Salas Fernandez and Roger Schaer and Abdel Aziz Taha and Marianne Winterstein and Allan Hanbury},
title = {VISCERAL - VISual Concept Extraction challenge in RAdioLogy : ISBI 2014 Challenge Organization},
booktitle = {VISCERAL Challenge at ISBI},
editor = {Orcun Goksel},
publisher = {CEUR-WS},
year = {2014},
number = {1194},
pages = {6-15},
url = {http://ceur-ws.org/Vol-1194/visceralISBI14-0.pdf}
}
Edited Books:
 [2021] Alfred Franz and Orcun Goksel:
"(eds.) IJCARS-IPCAI 2021 Special Issue: Proceedings of Information Processing for Computer-Assisted Interventions",
, 2021.
[2021] Alfred Franz and Orcun Goksel:
"(eds.) IJCARS-IPCAI 2021 Special Issue: Proceedings of Information Processing for Computer-Assisted Interventions",
, 2021.
@book{Franz_ipcai_21,
author = {Alfred Franz and Orcun Goksel},
title = {(eds.) IJCARS-IPCAI 2021 Special Issue: Proceedings of Information Processing for Computer-Assisted Interventions},
publisher = {Springer},
year = {2021},
doi = {10.1007/s11548-021-02401-5}
}
 [2020] Z. Spiclin, J. McClelland, J. Kybic, and O. Goksel:
"(eds.) Proceedings of International Workshop on Biomedical Image Registration (WBIR)",
, Dec 2020.
[2020] Z. Spiclin, J. McClelland, J. Kybic, and O. Goksel:
"(eds.) Proceedings of International Workshop on Biomedical Image Registration (WBIR)",
, Dec 2020.
@book{Spiclin_wbir_20,
author = {Spiclin, Z. and McClelland, J. and Kybic, J. and Goksel, O},
title = {(eds.) Proceedings of International Workshop on Biomedical Image Registration (WBIR)},
publisher = {Springer},
year = {2020},
doi = {10.1007/978-3-030-50120-4}
}
 [2018] A. Gooya, O. Goksel, I. Oguz, and N. Burgos:
"(eds.) Proceedings of Simulation and Synthesis in Medical Imaging (SASHIMI)",
, Sep 2018.
[2018] A. Gooya, O. Goksel, I. Oguz, and N. Burgos:
"(eds.) Proceedings of Simulation and Synthesis in Medical Imaging (SASHIMI)",
, Sep 2018.
@book{Gooya_sashimi_18,
author = {Gooya, A. and Goksel, O. and Oguz, I. and Burgos, N.},
title = {(eds.) Proceedings of Simulation and Synthesis in Medical Imaging (SASHIMI)},
publisher = {Springer},
year = {2018},
doi = {10.1007/978-3-030-00536-8}
}
 [2015] Orcun Goksel, Oscar Alfonso Jiménez del Toro, Antonio Foncubierta, and Henning Müller:
"(eds.) Proceedings of the VISCERAL Challenge at ISBI",
(1390), Apr 2015.
[2015] Orcun Goksel, Oscar Alfonso Jiménez del Toro, Antonio Foncubierta, and Henning Müller:
"(eds.) Proceedings of the VISCERAL Challenge at ISBI",
(1390), Apr 2015.
@book{Goksel_visceral_15,
author = {Orcun Goksel and Jim\'enez del Toro, Oscar Alfonso and Antonio Foncubierta and Henning M\"uller},
title = {(eds.) Proceedings of the VISCERAL Challenge at ISBI},
publisher = {CEUR-WS.org},
year = {2015},
number = {1390},
url = {http://ceur-ws.org/Vol-1390/}
}
@book{Goksel_visceral_14,
author = {Orcun Goksel},
title = {(ed.) Proceedings of the VISCERAL Challenge at ISBI},
publisher = {CEUR-WS.org},
year = {2014},
number = {1194},
url = {http://ceur-ws.org/Vol-1194}
}
Conference Contributions:
 [2023] Can Deniz Bezek and Orcun Goksel:
"Pulse-Echo Speed-of-Sound As Imaging Biomarker for Breast Density: Virtual Source Acquisitions for In-Vivo Application",
In IEEE Int Ultrasonics Symposium (IUS), Montreal, QC, Canada, Sep 2023.
[2023] Can Deniz Bezek and Orcun Goksel:
"Pulse-Echo Speed-of-Sound As Imaging Biomarker for Breast Density: Virtual Source Acquisitions for In-Vivo Application",
In IEEE Int Ultrasonics Symposium (IUS), Montreal, QC, Canada, Sep 2023.
@inproceedings{Bezek_pulse-echo_23,
author = {Can Deniz Bezek and Orcun Goksel},
title = {Pulse-Echo Speed-of-Sound As Imaging Biomarker for Breast Density: Virtual Source Acquisitions for In-Vivo Application},
booktitle = {IEEE Int Ultrasonics Symposium (IUS)},
year = {2023},
number = {2166}
}
 [2023] Can Deniz Bezek and Orcun Goksel:
"Sound-Speed Reconstruction with Learned Kernels Based on a Convolutional Formulation of Sound-Speed and Speckle-Shift Relation",
In IEEE Int Ultrasonics Symposium (IUS), Montreal, QC, Canada, Sep 2023.
[2023] Can Deniz Bezek and Orcun Goksel:
"Sound-Speed Reconstruction with Learned Kernels Based on a Convolutional Formulation of Sound-Speed and Speckle-Shift Relation",
In IEEE Int Ultrasonics Symposium (IUS), Montreal, QC, Canada, Sep 2023.
@inproceedings{Bezek_sound-speed_23,
author = {Can Deniz Bezek and Orcun Goksel},
title = {Sound-Speed Reconstruction with Learned Kernels Based on a Convolutional Formulation of Sound-Speed and Speckle-Shift Relation},
booktitle = {IEEE Int Ultrasonics Symposium (IUS)},
year = {2023},
number = {2144}
}
 [2023] Dieter Schweizer, Can Deniz Bezek, Monika Farkas, and Orcun Goksel:
"Motion Sensitivity of Transmit Sequences for Pulse-Echo Mapping of Sound Speed Based on Apparent Speckle Shifts",
In IEEE Int Ultrasonics Symposium (IUS), Montreal, QC, Canada, Sep 2023.
[2023] Dieter Schweizer, Can Deniz Bezek, Monika Farkas, and Orcun Goksel:
"Motion Sensitivity of Transmit Sequences for Pulse-Echo Mapping of Sound Speed Based on Apparent Speckle Shifts",
In IEEE Int Ultrasonics Symposium (IUS), Montreal, QC, Canada, Sep 2023.
@inproceedings{Schweizer_motion_23,
author = {Dieter Schweizer and Can Deniz Bezek and Monika Farkas and Orcun Goksel},
title = {Motion Sensitivity of Transmit Sequences for Pulse-Echo Mapping of Sound Speed Based on Apparent Speckle Shifts},
booktitle = {IEEE Int Ultrasonics Symposium (IUS)},
year = {2023},
number = {1970}
}
 [2023] Can Deniz Bezek, Lingkai Zhu, and Orcun Goksel:
"Model-based Deep Learning of Ultrasound Beamforming",
In Swedish Symposium on Image Analysis (SSBA), pp. 1-4, Kolmården, Sweden, Mar 2023.
[2023] Can Deniz Bezek, Lingkai Zhu, and Orcun Goksel:
"Model-based Deep Learning of Ultrasound Beamforming",
In Swedish Symposium on Image Analysis (SSBA), pp. 1-4, Kolmården, Sweden, Mar 2023.
@inproceedings{Bezek_model-based_23,
author = {Can Deniz Bezek and Lingkai Zhu and Orcun Goksel},
title = {Model-based Deep Learning of Ultrasound Beamforming},
booktitle = {Swedish Symposium on Image Analysis (SSBA)},
year = {2023},
pages = {1-4}
}
2022
 [2022] Kevin Thandiackal, Boqi Chen, Pushpak Pati, Guillaume Jaume, Drew F.K. Williamson, Maria Gabrani, and Orcun Goksel:
"Differentiable Zooming for Multiple Instance Learning on Whole-Slide Images",
In European Conference on Computer Vision (ECCV), Tel Aviv, Israel, Oct 2022.
[2022] Kevin Thandiackal, Boqi Chen, Pushpak Pati, Guillaume Jaume, Drew F.K. Williamson, Maria Gabrani, and Orcun Goksel:
"Differentiable Zooming for Multiple Instance Learning on Whole-Slide Images",
In European Conference on Computer Vision (ECCV), Tel Aviv, Israel, Oct 2022.
@inproceedings{Thandiackal_differentiable_22,
author = {Kevin Thandiackal and Boqi Chen and Pushpak Pati and Guillaume Jaume and Drew FK Williamson and Maria Gabrani and Orcun Goksel},
title = {Differentiable Zooming for Multiple Instance Learning on Whole-Slide Images},
booktitle = {European Conference on Computer Vision (ECCV)},
year = {2022},
url = {https://arxiv.org/abs/2204.12454},
doi = {10.1007/978-3-031-19803-8_41}
}
 [2022] Can Deniz Bezek, Mert Bilgin, Lin Zhang, and Orcun Goksel:
"Global Speed-of-Sound Prediction Using Transmission Geometry",
In IEEE Int Ultrasonics Symposium (IUS), Venice, Italy, Oct 2022.
[2022] Can Deniz Bezek, Mert Bilgin, Lin Zhang, and Orcun Goksel:
"Global Speed-of-Sound Prediction Using Transmission Geometry",
In IEEE Int Ultrasonics Symposium (IUS), Venice, Italy, Oct 2022.
@inproceedings{Bezek_global_22,
author = {Can Deniz Bezek and Mert Bilgin and Lin Zhang and Orcun Goksel},
title = {Global Speed-of-Sound Prediction Using Transmission Geometry},
booktitle = {IEEE Int Ultrasonics Symposium (IUS)},
year = {2022},
number = {arxiv:2208.08377},
url = {https://arxiv.org/abs/2208.08377}
}
 [2022] Alvaro Gomariz, Huanxiang Lu, Yun Yvonna Li, Thomas Albrecht, Andreas Maunz, Fethallah Benmansour, Alessandra M. Valcarcel, Jennifer Luu, Daniela Ferrara, and Orcun Goksel:
"Unsupervised Domain Adaptation with Contrastive Learning for OCT Segmentation",
In MICCAI, Singapore, Sep 2022.
[2022] Alvaro Gomariz, Huanxiang Lu, Yun Yvonna Li, Thomas Albrecht, Andreas Maunz, Fethallah Benmansour, Alessandra M. Valcarcel, Jennifer Luu, Daniela Ferrara, and Orcun Goksel:
"Unsupervised Domain Adaptation with Contrastive Learning for OCT Segmentation",
In MICCAI, Singapore, Sep 2022.
@inproceedings{Gomariz_unsupervised_22,
author = {Alvaro Gomariz and Huanxiang Lu and Yun Yvonna Li and Thomas Albrecht and Andreas Maunz and Fethallah Benmansour and Alessandra M Valcarcel and Jennifer Luu and Daniela Ferrara and Orcun Goksel},
title = {Unsupervised Domain Adaptation with Contrastive Learning for OCT Segmentation},
booktitle = {MICCAI},
year = {2022},
url = {https://arxiv.org/abs/2203.03664},
doi = {10.1007/978-3-031-16452-1_34}
}
 [2022] Alvaro Gomariz, Huanxiang Lu, Yun Li, Thomas Albrecht, Andreas Maunz, Fethallah Benmansour, Jennifer Luu, Orcun Goksel, and Daniela Ferrara:
"A unified deep learning approach for OCT segmentation from different devices and retinal diseases",
In ARVO Annual Meeting Procs in Investigative Ophthalmology and Visual Science, pp. 2053-F0042, Denver, USA, Jun 2022.
[2022] Alvaro Gomariz, Huanxiang Lu, Yun Li, Thomas Albrecht, Andreas Maunz, Fethallah Benmansour, Jennifer Luu, Orcun Goksel, and Daniela Ferrara:
"A unified deep learning approach for OCT segmentation from different devices and retinal diseases",
In ARVO Annual Meeting Procs in Investigative Ophthalmology and Visual Science, pp. 2053-F0042, Denver, USA, Jun 2022.
@inproceedings{Gomariz_unified_22,
author = {Alvaro Gomariz and Huanxiang Lu and Yun Li and Thomas Albrecht and Andreas Maunz and Fethallah Benmansour and Jennifer Luu and Orcun Goksel and Daniela Ferrara},
title = {A unified deep learning approach for OCT segmentation from different devices and retinal diseases},
booktitle = {ARVO Annual Meeting Procs in Investigative Ophthalmology and Visual Science},
year = {2022},
volume = {63},
number = {7},
pages = {2053-F0042},
url = {https://iovs.arvojournals.org/article.aspx?articleid=2782139}
}
 [2022] Can Deniz Bezek, Mert Bilgin, Lin Zhang, and Orcun Goksel:
"Mean Speed-of-Sound Estimation Using Geometric Disparities",
In Swedish Symposium on Image Analysis (SSBA), pp. 1-4, Uppsala, Sweden, Mar 2022.
[2022] Can Deniz Bezek, Mert Bilgin, Lin Zhang, and Orcun Goksel:
"Mean Speed-of-Sound Estimation Using Geometric Disparities",
In Swedish Symposium on Image Analysis (SSBA), pp. 1-4, Uppsala, Sweden, Mar 2022.
@inproceedings{Bezek_mean_22,
author = {Can Deniz Bezek and Mert Bilgin and Lin Zhang and Orcun Goksel},
title = {Mean Speed-of-Sound Estimation Using Geometric Disparities},
booktitle = {Swedish Symposium on Image Analysis (SSBA)},
year = {2022},
pages = {1-4}
}
2021
 [2021] Devavrat Tomar, Lin Zhang, Tiziano Portenier, and Orcun Goksel:
"Content-Preserving Unpaired Translation from Simulated to Realistic Ultrasound Images",
In MICCAI, Sep 2021.
[2021] Devavrat Tomar, Lin Zhang, Tiziano Portenier, and Orcun Goksel:
"Content-Preserving Unpaired Translation from Simulated to Realistic Ultrasound Images",
In MICCAI, Sep 2021.
@inproceedings{Tomar_content-preserving_21,
author = {Devavrat Tomar and Lin Zhang and Tiziano Portenier and Orcun Goksel},
title = {Content-Preserving Unpaired Translation from Simulated to Realistic Ultrasound Images},
booktitle = {MICCAI},
year = {2021},
url = {https://arxiv.org/abs/2103.05745},
doi = {10.1007/978-3-030-87237-3_63}
}
 [2021] Valentin Anklin, Pushpak Pati, Guillaume Jaume, Behzad Bozorgtabar, Antonio Foncubierta-Rodriguez, Jean-Philippe Thiran, Mathilde Sibony, Maria Gabrani, and Orcun Goksel:
"Learning Whole-Slide Segmentation from Inexact and Incomplete Labels using Tissue Graphs",
In MICCAI, pp. 636-646, Sep 2021.
[2021] Valentin Anklin, Pushpak Pati, Guillaume Jaume, Behzad Bozorgtabar, Antonio Foncubierta-Rodriguez, Jean-Philippe Thiran, Mathilde Sibony, Maria Gabrani, and Orcun Goksel:
"Learning Whole-Slide Segmentation from Inexact and Incomplete Labels using Tissue Graphs",
In MICCAI, pp. 636-646, Sep 2021.
@inproceedings{Anklin_learning_21,
author = {Valentin Anklin and Pushpak Pati and Guillaume Jaume and Behzad Bozorgtabar and Antonio Foncubierta-Rodriguez and Jean-Philippe Thiran and Mathilde Sibony and Maria Gabrani and Orcun Goksel},
title = {Learning Whole-Slide Segmentation from Inexact and Incomplete Labels using Tissue Graphs},
booktitle = {MICCAI},
year = {2021},
pages = {636-646},
url = {https://arxiv.org/abs/2103.03129},
doi = {10.1007/978-3-030-87196-3_59}
}
 [2021] Xenia Augustin, Lin Zhang, and Orcun Goksel:
"Estimating Mean Speed-of-Sound from Sequence-Dependent Geometric Disparities",
In IEEE Int Ultrasonics Symposium (IUS), pp. 1-4, Sep 2021.
[2021] Xenia Augustin, Lin Zhang, and Orcun Goksel:
"Estimating Mean Speed-of-Sound from Sequence-Dependent Geometric Disparities",
In IEEE Int Ultrasonics Symposium (IUS), pp. 1-4, Sep 2021.
@inproceedings{Augustin_estimating_21,
author = {Xenia Augustin and Lin Zhang and Orcun Goksel},
title = {Estimating Mean Speed-of-Sound from Sequence-Dependent Geometric Disparities},
booktitle = {IEEE Int Ultrasonics Symposium (IUS)},
year = {2021},
pages = {1-4},
url = {https://arxiv.org/abs/2109.11819},
doi = {10.1109/IUS52206.2021.9593742}
}
 [2021] Guillaume Jaume, Pushpak Pati, Behzad Bozorgtabar, Antonio Foncubierta-Rodríguez, Florinda Feroce, Anna Maria Anniciello, Tilman Rau, Jean-Philippe Thiran, Maria Gabrani, and Orcun Goksel:
"Quantifying Explainers of Graph Neural Networks in Computational Pathology",
In Computer Vision and Pattern Recognition (CVPR), pp. 8102-12, Jun 2021.
[2021] Guillaume Jaume, Pushpak Pati, Behzad Bozorgtabar, Antonio Foncubierta-Rodríguez, Florinda Feroce, Anna Maria Anniciello, Tilman Rau, Jean-Philippe Thiran, Maria Gabrani, and Orcun Goksel:
"Quantifying Explainers of Graph Neural Networks in Computational Pathology",
In Computer Vision and Pattern Recognition (CVPR), pp. 8102-12, Jun 2021.
@inproceedings{Jaume_quantifying_21,
author = {Guillaume Jaume and Pushpak Pati and Behzad Bozorgtabar and Antonio Foncubierta-Rodr\'iguez and Florinda Feroce and Anna Maria Anniciello and Tilman Rau and Jean-Philippe Thiran and Maria Gabrani and Orcun Goksel},
title = {Quantifying Explainers of Graph Neural Networks in Computational Pathology},
booktitle = {Computer Vision and Pattern Recognition (CVPR)},
year = {2021},
pages = {8102-12},
url = {https://arxiv.org/abs/2011.12646},
doi = {10.1109/CVPR46437.2021.00801}
}
 [2021] Alvaro Gomariz, Raphael Egli, Tiziano Portenier, César Nombela-Arrieta, and Orcun Goksel:
"Utilizing Uncertainty Estimation in Deep Learning Segmentation of Fluorescence Microscopy Images with Missing Markers",
In IEEE Int Symposium on Biomedical Imaging (ISBI), pp. 371-4, Apr 2021.
[2021] Alvaro Gomariz, Raphael Egli, Tiziano Portenier, César Nombela-Arrieta, and Orcun Goksel:
"Utilizing Uncertainty Estimation in Deep Learning Segmentation of Fluorescence Microscopy Images with Missing Markers",
In IEEE Int Symposium on Biomedical Imaging (ISBI), pp. 371-4, Apr 2021.
@inproceedings{Gomariz_utilizing_21,
author = {Alvaro Gomariz and Raphael Egli and Tiziano Portenier and C\'esar Nombela-Arrieta and Orcun Goksel},
title = {Utilizing Uncertainty Estimation in Deep Learning Segmentation of Fluorescence Microscopy Images with Missing Markers},
booktitle = {IEEE Int Symposium on Biomedical Imaging (ISBI)},
year = {2021},
pages = {371-4},
url = {https://arxiv.org/abs/2101.11476},
doi = {10.1109/ISBI48211.2021.9434158}
}
 [2021] Bhaskara Rao Chintada, Richard Rau, and Orcun Goksel:
"Time of Arrival Delineation In Echo Traces For Reflection Ultrasound Tomography",
In IEEE Int Symposium on Biomedical Imaging (ISBI), pp. 1342-5, Apr 2021.
[2021] Bhaskara Rao Chintada, Richard Rau, and Orcun Goksel:
"Time of Arrival Delineation In Echo Traces For Reflection Ultrasound Tomography",
In IEEE Int Symposium on Biomedical Imaging (ISBI), pp. 1342-5, Apr 2021.
@inproceedings{Chintada_time_21,
author = {Bhaskara Rao Chintada and Richard Rau and Orcun Goksel},
title = {Time of Arrival Delineation In Echo Traces For Reflection Ultrasound Tomography},
booktitle = {IEEE Int Symposium on Biomedical Imaging (ISBI)},
year = {2021},
pages = {1342-5},
doi = {10.1109/ISBI48211.2021.9433846}
}
2020
 [2020] Tiziano Portenier, Siavash Bigdeli, and Orcun Goksel:
"GramGAN: Deep 3D Texture Synthesis From 2D Exemplars",
In Neural Information Processing Systems (NeurIPS), pp. 1-11, Dec 2020.
[2020] Tiziano Portenier, Siavash Bigdeli, and Orcun Goksel:
"GramGAN: Deep 3D Texture Synthesis From 2D Exemplars",
In Neural Information Processing Systems (NeurIPS), pp. 1-11, Dec 2020.
@inproceedings{Portenier_gramgan_20,
author = {Tiziano Portenier and Siavash Bigdeli and Orcun Goksel},
title = {GramGAN: Deep 3D Texture Synthesis From 2D Exemplars},
booktitle = {Neural Information Processing Systems (NeurIPS)},
year = {2020},
pages = {1-11},
url = {https://proceedings.neurips.cc/paper_files/paper/2020/hash/4df5bde009073d3ef60da64d736724d6-Abstract.html}
}
 [2020] Pushpak Pati, Guillaume Jaume, Lauren Alisha Fernandes, Antonio Foncubierta-Rodriguez, Florinda Feroce, Anna Maria Anniciello, Giosue Scognamiglio, Nadia Brancati, Daniel Riccio, Maurizio Di Bonito, Giuseppe De Pietro, Gerardo Botti, Orcun Goksel, Jean-Philippe Thiran, Maria Frucci, and Maria Gabrani:
"HACT-Net: A Hierarchical Cell-to-Tissue Graph Neural Network for Histopathological Image Classification",
In MICCAI Workshop on Graphs in Biomedical Image Analysis (GRAIL), pp. 208-219, Oct 2020.
[2020] Pushpak Pati, Guillaume Jaume, Lauren Alisha Fernandes, Antonio Foncubierta-Rodriguez, Florinda Feroce, Anna Maria Anniciello, Giosue Scognamiglio, Nadia Brancati, Daniel Riccio, Maurizio Di Bonito, Giuseppe De Pietro, Gerardo Botti, Orcun Goksel, Jean-Philippe Thiran, Maria Frucci, and Maria Gabrani:
"HACT-Net: A Hierarchical Cell-to-Tissue Graph Neural Network for Histopathological Image Classification",
In MICCAI Workshop on Graphs in Biomedical Image Analysis (GRAIL), pp. 208-219, Oct 2020.
* Best Paper Award
@inproceedings{Pati_hact-net_20,
author = {Pushpak Pati and Guillaume Jaume and Lauren Alisha Fernandes and Antonio Foncubierta-Rodriguez and Florinda Feroce and Anna Maria Anniciello and Giosue Scognamiglio and Nadia Brancati and Daniel Riccio and Maurizio Di Bonito and Giuseppe De Pietro and Gerardo Botti and Orcun Goksel and Jean-Philippe Thiran and Maria Frucci and Maria Gabrani},
title = {HACT-Net: A Hierarchical Cell-to-Tissue Graph Neural Network for Histopathological Image Classification},
booktitle = {MICCAI Workshop on Graphs in Biomedical Image Analysis (GRAIL)},
year = {2020},
pages = {208-219},
url = {https://arxiv.org/abs/2007.00584},
doi = {10.1007/978-3-030-60365-6_20}
}
 [2020] Emanuel Joos, Fabien Péan, and Orcun Goksel:
"Reinforcement Learning of Musculoskeletal Control from Functional Simulations",
In MICCAI, pp. 135-145, Oct 2020.
[2020] Emanuel Joos, Fabien Péan, and Orcun Goksel:
"Reinforcement Learning of Musculoskeletal Control from Functional Simulations",
In MICCAI, pp. 135-145, Oct 2020.
@inproceedings{Joos_recurrent_20,
author = {Emanuel Joos and Fabien P\'ean and Orcun Goksel},
title = {Reinforcement Learning of Musculoskeletal Control from Functional Simulations},
booktitle = {MICCAI},
year = {2020},
pages = {135-145},
url = {https://arxiv.org/abs/2007.06669},
doi = {10.1007/978-3-030-59716-0_14}
}
 [2020] Lin Zhang, Tiziano Portenier, Christoph Paulus, and Orcun Goksel:
"Deep Image Translation for Enhancing Simulated Ultrasound Images",
In MICCAI Workshop on Advances in Simplifying Medical Ultrasound, pp. 85-94, Oct 2020.
[2020] Lin Zhang, Tiziano Portenier, Christoph Paulus, and Orcun Goksel:
"Deep Image Translation for Enhancing Simulated Ultrasound Images",
In MICCAI Workshop on Advances in Simplifying Medical Ultrasound, pp. 85-94, Oct 2020.
@inproceedings{Zhang_deepI_20,
author = {Lin Zhang and Tiziano Portenier and Christoph Paulus and Orcun Goksel},
title = {Deep Image Translation for Enhancing Simulated Ultrasound Images},
booktitle = {MICCAI Workshop on Advances in Simplifying Medical Ultrasound},
year = {2020},
pages = {85-94},
url = {https://arxiv.org/abs/2006.10850},
doi = {10.1007/978-3-030-60334-2_9}
}
 [2020] Richard Rau, Ece Ozkan, Batu M. Ozturkler, Leila Gastli, and Orcun Goksel:
"Displacement Estimation Methods for Speed-of-Sound Imaging in Pulse-Echo",
In IEEE Int Ultrasonics Symposium (IUS), pp. 2007-2010, Sep 2020.
[2020] Richard Rau, Ece Ozkan, Batu M. Ozturkler, Leila Gastli, and Orcun Goksel:
"Displacement Estimation Methods for Speed-of-Sound Imaging in Pulse-Echo",
In IEEE Int Ultrasonics Symposium (IUS), pp. 2007-2010, Sep 2020.
@inproceedings{Rau_displacement_20,
author = {Richard Rau and Ece Ozkan and Batu M. Ozturkler and Leila Gastli and Orcun Goksel},
title = {Displacement Estimation Methods for Speed-of-Sound Imaging in Pulse-Echo},
booktitle = {IEEE Int Ultrasonics Symposium (IUS)},
year = {2020},
pages = {2007-2010},
doi = {10.1109/IUS46767.2020.9251781}
}
 [2020] Richard Rau, Justine Robin, Berkan Lafci, Aileen Schroeter, Michael Reiss, X. Luis Dean Ben, Orcun Goksel, and Daniel Razansky:
"Functional Neuroimaging of Mice Using Ultrasound and Optoacoustics",
In IEEE Int Ultrasonics Symposium (IUS), Sep 2020.
[2020] Richard Rau, Justine Robin, Berkan Lafci, Aileen Schroeter, Michael Reiss, X. Luis Dean Ben, Orcun Goksel, and Daniel Razansky:
"Functional Neuroimaging of Mice Using Ultrasound and Optoacoustics",
In IEEE Int Ultrasonics Symposium (IUS), Sep 2020.
@inproceedings{Rau_functional_20,
author = {Richard Rau and Justine Robin and Berkan Lafci and Aileen Schroeter and Michael Reiss and X Luis Dean Ben and Orcun Goksel and Daniel Razansky},
title = {Functional Neuroimaging of Mice Using Ultrasound and Optoacoustics},
booktitle = {IEEE Int Ultrasonics Symposium (IUS)},
year = {2020}
}
 [2020] Bhaskara Rao Chintada, Richard Rau, and Orcun Goksel:
"Model-Independent Quantification of Complex Shear Modulus via Speed and Attenuation of Surface Waves",
In IEEE Int Ultrasonics Symposium (IUS), pp. 1269-1272, Sep 2020.
[2020] Bhaskara Rao Chintada, Richard Rau, and Orcun Goksel:
"Model-Independent Quantification of Complex Shear Modulus via Speed and Attenuation of Surface Waves",
In IEEE Int Ultrasonics Symposium (IUS), pp. 1269-1272, Sep 2020.
@inproceedings{Chintada_model-independent_20,
author = {Bhaskara Rao Chintada and Richard Rau and Orcun Goksel},
title = {Model-Independent Quantification of Complex Shear Modulus via Speed and Attenuation of Surface Waves},
booktitle = {IEEE Int Ultrasonics Symposium (IUS)},
year = {2020},
pages = {1269-1272},
url = {pub/Chintada_model-independent_20.pdf},
doi = {10.1109/IUS46767.2020.9251721}
}
 [2020] Guillaume Jaume, Pushpak Pati, Antonio Foncubierta-Rodriguez, Florinda Feroce, Giosue Scognamiglio, Anna Maria Anniciello, Jean-Philippe Thiran, Orcun Goksel, and Maria Gabrani:
"Towards Explainable Graph Representations in Digital Pathology",
In ICML Workshop on Computational Biology, pp. 1-5, Jul 2020.
[2020] Guillaume Jaume, Pushpak Pati, Antonio Foncubierta-Rodriguez, Florinda Feroce, Giosue Scognamiglio, Anna Maria Anniciello, Jean-Philippe Thiran, Orcun Goksel, and Maria Gabrani:
"Towards Explainable Graph Representations in Digital Pathology",
In ICML Workshop on Computational Biology, pp. 1-5, Jul 2020.
@inproceedings{Jaume_towards_20,
author = {Guillaume Jaume and Pushpak Pati and Antonio Foncubierta-Rodriguez and Florinda Feroce and Giosue Scognamiglio and Anna Maria Anniciello and Jean-Philippe Thiran and Orcun Goksel and Maria Gabrani},
title = {Towards Explainable Graph Representations in Digital Pathology},
booktitle = {ICML Workshop on Computational Biology},
year = {2020},
pages = {1-5},
url = {https://arxiv.org/abs/2007.00311}
}
 [2020] Pushpak Pati, Antonio Foncubierta-Rodríguez, Orcun Goksel, and Maria Gabrani:
"Mitosis Detection under Limited Annotation: A Joint Learning Approach",
In IEEE Int Symp Biomedical Imaging (ISBI), pp. 486-489, Iowa City, USA, Apr 2020.
[2020] Pushpak Pati, Antonio Foncubierta-Rodríguez, Orcun Goksel, and Maria Gabrani:
"Mitosis Detection under Limited Annotation: A Joint Learning Approach",
In IEEE Int Symp Biomedical Imaging (ISBI), pp. 486-489, Iowa City, USA, Apr 2020.
@inproceedings{Pati_mitosis_20,
author = {Pushpak Pati and Antonio Foncubierta-Rodr\'{i}guez and Orcun Goksel and Maria Gabrani},
title = {Mitosis Detection under Limited Annotation: A Joint Learning Approach},
booktitle = {IEEE Int Symp Biomedical Imaging (ISBI)},
year = {2020},
pages = {486-489},
url = {https://arxiv.org/abs/2006.09772},
doi = {10.1109/ISBI45749.2020.9098431}
}
2019
 [2019] Richard Rau, Ozan Unal, Dieter Schweizer, Valery Vishnevskiy, and Orcun Goksel:
"Attenuation Imaging with Pulse-Echo Ultrasound based on an Acoustic Reflector",
In MICCAI, pp. 601-609, Shenzhen, China, Oct 2019.
[2019] Richard Rau, Ozan Unal, Dieter Schweizer, Valery Vishnevskiy, and Orcun Goksel:
"Attenuation Imaging with Pulse-Echo Ultrasound based on an Acoustic Reflector",
In MICCAI, pp. 601-609, Shenzhen, China, Oct 2019.
* Received MICCAI Best Presentation Award (1730 paper submissions in MICCAI 2019)
@inproceedings{Rau_attenuation_19,
author = {Rau, Richard and Ozan Unal and Dieter Schweizer and Valery Vishnevskiy and Orcun Goksel},
title = {Attenuation Imaging with Pulse-Echo Ultrasound based on an Acoustic Reflector},
booktitle = {MICCAI},
year = {2019},
pages = {601-609},
url = {https://arxiv.org/abs/1906.11615},
doi = {10.1007/978-3-030-32254-0_67}
}
 [2019] Valery Vishnevskiy, Richard Rau, and Orcun Goksel:
"Deep Variational Networks with Exponential Weighting for Learning Computed Tomography",
In MICCAI, pp. 310-318, Shenzhen, China, Oct 2019.
[2019] Valery Vishnevskiy, Richard Rau, and Orcun Goksel:
"Deep Variational Networks with Exponential Weighting for Learning Computed Tomography",
In MICCAI, pp. 310-318, Shenzhen, China, Oct 2019.
* Among 15 Finalists for MICCAI Best Presentation Award (1730 paper submissions to MICCAI 2019)
@inproceedings{Vishnevskiy_deep_19,
author = {Valery Vishnevskiy and Richard Rau and Orcun Goksel},
title = {Deep Variational Networks with Exponential Weighting for Learning Computed Tomography},
booktitle = {MICCAI},
year = {2019},
pages = {310-318},
url = {https://arxiv.org/abs/1906.05528},
doi = {10.1007/978-3-030-32226-7_35}
}
 [2019] Richard Rau, Dieter Schweizer, Valery Vishnevskiy, and Orcun Goksel:
"Ultrasound Aberration Correction based on Local Speed-of-Sound Map Estimation",
In IEEE Int Ultrasonics Symposium (IUS), pp. 1-4, Glasgow, Scotland, Oct 2019.
[2019] Richard Rau, Dieter Schweizer, Valery Vishnevskiy, and Orcun Goksel:
"Ultrasound Aberration Correction based on Local Speed-of-Sound Map Estimation",
In IEEE Int Ultrasonics Symposium (IUS), pp. 1-4, Glasgow, Scotland, Oct 2019.
@inproceedings{Rau_ultrasound_19,
author = {Richard Rau and Dieter Schweizer and Valery Vishnevskiy and Orcun Goksel},
title = {Ultrasound Aberration Correction based on Local Speed-of-Sound Map Estimation},
booktitle = {IEEE Int Ultrasonics Symposium (IUS)},
year = {2019},
pages = {1-4},
url = {https://arxiv.org/abs/1909.10254}
}
 [2019] Bhaskara Rao Chintada, Richard Rau, and Orcun Goksel:
"Acoustoelasticity analysis of shear waves for nonlinear biomechanical characterization of oil-gelatin phantoms",
In IEEE Int Ultrasonics Symposium (IUS), pp. 423-6, Glasgow, Scotland, Oct 2019.
[2019] Bhaskara Rao Chintada, Richard Rau, and Orcun Goksel:
"Acoustoelasticity analysis of shear waves for nonlinear biomechanical characterization of oil-gelatin phantoms",
In IEEE Int Ultrasonics Symposium (IUS), pp. 423-6, Glasgow, Scotland, Oct 2019.
@inproceedings{Chintada_acousto_19,
author = {Bhaskara Rao Chintada and Richard Rau and Orcun Goksel},
title = {Acoustoelasticity analysis of shear waves for nonlinear biomechanical characterization of oil-gelatin phantoms},
booktitle = {IEEE Int Ultrasonics Symposium (IUS)},
year = {2019},
pages = {423-6},
url = {https://www.research-collection.ethz.ch/handle/20.500.11850/403888},
doi = {10.1109/ULTSYM.2019.8925670}
}
 [2019] Andrawes Al Bahou, Christine Tanner, and Orcun Goksel:
"ScatGAN for Reconstruction of Ultrasound Scatterers using Generative Adversarial Networks",
In IEEE Int Symposium on Biomedical Imaging (ISBI), pp. 1674-7, Venice, Italy, Apr 2019.
[2019] Andrawes Al Bahou, Christine Tanner, and Orcun Goksel:
"ScatGAN for Reconstruction of Ultrasound Scatterers using Generative Adversarial Networks",
In IEEE Int Symposium on Biomedical Imaging (ISBI), pp. 1674-7, Venice, Italy, Apr 2019.
@inproceedings{AlBahou_scatgan_19,
author = {Al Bahou, Andrawes and Christine Tanner and Orcun Goksel},
title = {ScatGAN for Reconstruction of Ultrasound Scatterers using Generative Adversarial Networks},
booktitle = {IEEE Int Symposium on Biomedical Imaging (ISBI)},
year = {2019},
pages = {1674-7},
url = {https://arxiv.org/abs/1902.00469},
doi = {10.1109/ISBI.2019.8759251}
}
 [2019] Lin Zhang, Valery Vishnevsky, Andras Jakab, and Orcun Goksel:
"Implicit modeling with uncertainty estimation for intravoxel incoherent motion imaging",
In IEEE Int Symposium on Biomedical Imaging (ISBI), pp. 1003-7, Venice, Italy, Apr 2019.
[2019] Lin Zhang, Valery Vishnevsky, Andras Jakab, and Orcun Goksel:
"Implicit modeling with uncertainty estimation for intravoxel incoherent motion imaging",
In IEEE Int Symposium on Biomedical Imaging (ISBI), pp. 1003-7, Venice, Italy, Apr 2019.
@inproceedings{Zhang_implicit_19,
author = {Lin Zhang and Valery Vishnevsky and Andras Jakab and Orcun Goksel},
title = {Implicit modeling with uncertainty estimation for intravoxel incoherent motion imaging},
booktitle = {IEEE Int Symposium on Biomedical Imaging (ISBI)},
year = {2019},
pages = {1003-7},
url = {https://arxiv.org/abs/1810.10358},
doi = {10.1109/ISBI.2019.8759391}
}
 [2019] Alvaro Gomariz, Weiye Li, Ece Ozkan, Christine Tanner, and Orcun Goksel:
"Siamese Networks with Location Prior for Landmark Tracking in Liver Ultrasound Sequences",
In IEEE Int Symposium on Biomedical Imaging (ISBI), pp. 1757-60, Venice, Italy, Apr 2019.
[2019] Alvaro Gomariz, Weiye Li, Ece Ozkan, Christine Tanner, and Orcun Goksel:
"Siamese Networks with Location Prior for Landmark Tracking in Liver Ultrasound Sequences",
In IEEE Int Symposium on Biomedical Imaging (ISBI), pp. 1757-60, Venice, Italy, Apr 2019.
@inproceedings{Gomariz_siamese_19,
author = {Alvaro Gomariz and Weiye Li and Ece Ozkan and Christine Tanner and Orcun Goksel},
title = {Siamese Networks with Location Prior for Landmark Tracking in Liver Ultrasound Sequences},
booktitle = {IEEE Int Symposium on Biomedical Imaging (ISBI)},
year = {2019},
pages = {1757-60},
url = {https://arxiv.org/abs/1901.08109},
doi = {10.1109/ISBI.2019.8759382}
}
 [2019] Matija Ciganovic, Firat Ozdemir, Mazda Farshad, and Orcun Goksel:
"Deep learning techniques for bone surface delineation in ultrasound",
In SPIE Medical Imaging, pp. 10955-33, San Diego, CA, USA, Feb 2019.
[2019] Matija Ciganovic, Firat Ozdemir, Mazda Farshad, and Orcun Goksel:
"Deep learning techniques for bone surface delineation in ultrasound",
In SPIE Medical Imaging, pp. 10955-33, San Diego, CA, USA, Feb 2019.
@inproceedings{Ciganovic_deep_19,
author = {Matija Ciganovic and Firat Ozdemir and Mazda Farshad and Orcun Goksel},
title = {Deep learning techniques for bone surface delineation in ultrasound},
booktitle = {SPIE Medical Imaging},
year = {2019},
pages = {10955-33},
url = {https://www.research-collection.ethz.ch/handle/20.500.11850/350278},
doi = {10.1117/12.2512997}
}
 [2019] Pushpak Pati, Raul Catena, Orcun Goksel, and Maria Gabrani:
"A deep learning framework for context-aware mitotic activity estimation in whole slide images",
In SPIE Medical Imaging, pp. 10956-09, San Diego, CA, USA, Feb 2019.
[2019] Pushpak Pati, Raul Catena, Orcun Goksel, and Maria Gabrani:
"A deep learning framework for context-aware mitotic activity estimation in whole slide images",
In SPIE Medical Imaging, pp. 10956-09, San Diego, CA, USA, Feb 2019.
@inproceedings{Pati_deep_19,
author = {Pushpak Pati and Raul Catena and Orcun Goksel and Maria Gabrani},
title = {A deep learning framework for context-aware mitotic activity estimation in whole slide images},
booktitle = {SPIE Medical Imaging},
year = {2019},
pages = {10956-09},
doi = {10.1117/12.2512705}
}
2018
 [2018] Valery Vishnevskiy, Sergio J. Sanabria, and Orcun Goksel:
"Image Reconstruction via Variational Network for Real-Time Hand-Held Sound-Speed Imaging",
In MICCAI Workshop on Machine Learning for Medical Image Reconstruction, pp. 120-128, Granada, Spain, Sep 2018.
[2018] Valery Vishnevskiy, Sergio J. Sanabria, and Orcun Goksel:
"Image Reconstruction via Variational Network for Real-Time Hand-Held Sound-Speed Imaging",
In MICCAI Workshop on Machine Learning for Medical Image Reconstruction, pp. 120-128, Granada, Spain, Sep 2018.
@inproceedings{Vishnevskiy_image_18,
author = {Valery Vishnevskiy and Sergio J Sanabria and Orcun Goksel},
title = {Image Reconstruction via Variational Network for Real-Time Hand-Held Sound-Speed Imaging},
booktitle = {MICCAI Workshop on Machine Learning for Medical Image Reconstruction},
year = {2018},
pages = {120-128},
url = {https://arxiv.org/abs/1807.07416},
doi = {10.1007/978-3-030-00129-2_14}
}
 [2018] Firat Ozdemir, Philipp Fuernstahl, and Orcun Goksel:
"Learn the New, Keep the Old: Extending Pretrained Models with New Anatomy and Images",
In MICCAI, pp. 361-369, Granada, Spain, Sep 2018.
[2018] Firat Ozdemir, Philipp Fuernstahl, and Orcun Goksel:
"Learn the New, Keep the Old: Extending Pretrained Models with New Anatomy and Images",
In MICCAI, pp. 361-369, Granada, Spain, Sep 2018.
@inproceedings{Ozdemir_learn_18,
author = {Firat Ozdemir and Philipp Fuernstahl and Orcun Goksel},
title = {Learn the New, Keep the Old: Extending Pretrained Models with New Anatomy and Images},
booktitle = {MICCAI},
year = {2018},
pages = {361-369},
url = {https://arxiv.org/abs/1806.00265},
doi = {10.1007/978-3-030-00937-3_42}
}
 [2018] Christine Tanner, Rastislav Starkov, Michael Bajka, and Orcun Goksel:
"Framework for Fusion of Data- and Model-Based Approaches for Ultrasound Simulation",
In MICCAI, pp. 332-339, Granada, Spain, Sep 2018.
[2018] Christine Tanner, Rastislav Starkov, Michael Bajka, and Orcun Goksel:
"Framework for Fusion of Data- and Model-Based Approaches for Ultrasound Simulation",
In MICCAI, pp. 332-339, Granada, Spain, Sep 2018.
* also invited talk at 2018 Eurographic Workshop on Visual Computing for Biology and Medicine [VCBM]
@inproceedings{Tanner_combining_18,
author = {Christine Tanner and Rastislav Starkov and Michael Bajka and Orcun Goksel},
title = {Framework for Fusion of Data- and Model-Based Approaches for Ultrasound Simulation},
booktitle = {MICCAI},
year = {2018},
pages = {332-339},
doi = {10.1007/978-3-030-00937-3_39}
}
 [2018] Firat Ozdemir, Zixuan Peng, Christine Tanner, Philipp Fuernstahl, and Orcun Goksel:
"Active Learning for Segmentation by Optimizing Content Information for Maximal Entropy",
In MICCAI Workshop on Deep Learning in Medical Image Analysis, pp. 183-191, Granada, Spain, Sep 2018.
[2018] Firat Ozdemir, Zixuan Peng, Christine Tanner, Philipp Fuernstahl, and Orcun Goksel:
"Active Learning for Segmentation by Optimizing Content Information for Maximal Entropy",
In MICCAI Workshop on Deep Learning in Medical Image Analysis, pp. 183-191, Granada, Spain, Sep 2018.
@inproceedings{Ozdemir_active_18,
author = {Firat Ozdemir and Zixuan Peng and Christine Tanner and Philipp Fuernstahl and Orcun Goksel},
title = {Active Learning for Segmentation by Optimizing Content Information for Maximal Entropy},
booktitle = {MICCAI Workshop on Deep Learning in Medical Image Analysis},
year = {2018},
pages = {183-191},
url = {https://arxiv.org/abs/1807.07349},
doi = {10.1007/978-3-030-00889-5_21}
}
 [2018] Kevin Thandiackal and Orcun Goksel:
"A Structure-aware Convolutional Neural Network for Skin Lesion Classification",
In MICCAI Workshop on ISIC Skin Image Analysis, pp. 312-319, Granada, Spain, Sep 2018.
[2018] Kevin Thandiackal and Orcun Goksel:
"A Structure-aware Convolutional Neural Network for Skin Lesion Classification",
In MICCAI Workshop on ISIC Skin Image Analysis, pp. 312-319, Granada, Spain, Sep 2018.
@inproceedings{Thandiackal_structure-aware_18,
author = {Kevin Thandiackal and Orcun Goksel},
title = {A Structure-aware Convolutional Neural Network for Skin Lesion Classification},
booktitle = {MICCAI Workshop on ISIC Skin Image Analysis},
year = {2018},
pages = {312-319},
doi = {10.1007/978-3-030-01201-4_34}
}
 [2018] Corin F. Otesteanu, Valeriy Vishnevskiy, Christian Guenthner, Sebastian Kozerke, and Orcun Goksel:
"Ultrasound and Magnetic-Resonance Harmonic Elastography with Hybrid Inverse-Problem Formulation of FEM Viscoelasticity Reconstruction",
In Int Tissue Elasticity Conf (ITEC), Avignon, France, Sep 2018.
[2018] Corin F. Otesteanu, Valeriy Vishnevskiy, Christian Guenthner, Sebastian Kozerke, and Orcun Goksel:
"Ultrasound and Magnetic-Resonance Harmonic Elastography with Hybrid Inverse-Problem Formulation of FEM Viscoelasticity Reconstruction",
In Int Tissue Elasticity Conf (ITEC), Avignon, France, Sep 2018.
@inproceedings{Otesteanu_ultrasound_18,
author = {Corin F. Otesteanu and Valeriy Vishnevskiy and Christian Guenthner and Sebastian Kozerke and Orcun Goksel},
title = {Ultrasound and Magnetic-Resonance Harmonic Elastography with Hybrid Inverse-Problem Formulation of FEM Viscoelasticity Reconstruction},
booktitle = {Int Tissue Elasticity Conf (ITEC)},
year = {2018}
}
 [2018] Lisa Ruby, Sergio J. Sanabria, Anika S. Obrist, Katharina Martini, Serafino Forte, Orcun Goksel, Thomas Frauenfelder, Rahel Kubik-Huch, and Marga B. Rominger:
"Menstrual cycle-related changes in breast density using hand-held Speed-of-Sound Ultrasound",
In Dreiländertreffen Ultraschall, pp. S43-4, Basel, Switzerland, Nov 2018.
[2018] Lisa Ruby, Sergio J. Sanabria, Anika S. Obrist, Katharina Martini, Serafino Forte, Orcun Goksel, Thomas Frauenfelder, Rahel Kubik-Huch, and Marga B. Rominger:
"Menstrual cycle-related changes in breast density using hand-held Speed-of-Sound Ultrasound",
In Dreiländertreffen Ultraschall, pp. S43-4, Basel, Switzerland, Nov 2018.
@inproceedings{Ruby_menstrual_18,
author = {Lisa Ruby and Sergio J Sanabria and Anika S Obrist and Katharina Martini and Serafino Forte and Orcun Goksel and Thomas Frauenfelder and Rahel Kubik-Huch and Marga B Rominger},
title = {Menstrual cycle-related changes in breast density using hand-held Speed-of-Sound Ultrasound},
booktitle = {Dreiländertreffen Ultraschall},
journal = {Ultraschall in der Medizin},
year = {2018},
volume = {39},
pages = {S43-4},
doi = {10.1055/s-0038-1670479}
}
 [2018] A. Jakab, Z. Goey, O. Goksel, R. Tuura, R. Bauer, L. Stieglitz, G. Szekely, E. Martin, and B. Werner:
"NeuroShape: a novel software implementing state-of-the-art targeting for MRI-guided focused ultrasound neurosurgery",
In Swiss Congress of Radiology, May 2018.
[2018] A. Jakab, Z. Goey, O. Goksel, R. Tuura, R. Bauer, L. Stieglitz, G. Szekely, E. Martin, and B. Werner:
"NeuroShape: a novel software implementing state-of-the-art targeting for MRI-guided focused ultrasound neurosurgery",
In Swiss Congress of Radiology, May 2018.
@inproceedings{Jakab_neuroshape_18,
author = {A. Jakab and Z. Goey and O. Goksel and R. Tuura and R. Bauer and L. Stieglitz and G. Szekely and E. Martin and B. Werner},
title = {NeuroShape: a novel software implementing state-of-the-art targeting for MRI-guided focused ultrasound neurosurgery},
booktitle = {Swiss Congress of Radiology},
year = {2018},
doi = {10.13140/RG.2.2.16998.73283}
}
 [2018] Fabien Péan, Philipp Fŭrnstahl, and Orcun Goksel:
"A musculoskeletal model of the shoulder combining multibody dynamics and FEM using B-Spline elements",
In Int Symp Computer Methods in Biomechanics and Biomedical Engineering (CMBBE), Lisbon, Portugal, Mar 2018.
[2018] Fabien Péan, Philipp Fŭrnstahl, and Orcun Goksel:
"A musculoskeletal model of the shoulder combining multibody dynamics and FEM using B-Spline elements",
In Int Symp Computer Methods in Biomechanics and Biomedical Engineering (CMBBE), Lisbon, Portugal, Mar 2018.
@inproceedings{Pean_musculoskeletal_18,
author = {Fabien P\'{e}an and Philipp F\u{u}rnstahl and Orcun Goksel},
title = {A musculoskeletal model of the shoulder combining multibody dynamics and FEM using B-Spline elements},
booktitle = {Int Symp Computer Methods in Biomechanics and Biomedical Engineering (CMBBE)},
year = {2018},
url = {http://cmbbe2018.tecnico.ulisboa.pt/pen_cmbbe2018/pdf/WEB_ABSTRACTS/Abstracts_CMBBE2018_149.pdf}
}
 [2018] Sergio Sanabria, Katharina Martini, Marga Rominger, Konstantin Dedes, D. Vorburger, Thomas Frauenfelder, and Orcun Goksel:
"Breast cancer assessment with hand-held ultrasound based Speed of Sound: preliminary results",
In European Conference on Radiology (ECR), Vienna, Austria, Feb 2018.
[2018] Sergio Sanabria, Katharina Martini, Marga Rominger, Konstantin Dedes, D. Vorburger, Thomas Frauenfelder, and Orcun Goksel:
"Breast cancer assessment with hand-held ultrasound based Speed of Sound: preliminary results",
In European Conference on Radiology (ECR), Vienna, Austria, Feb 2018.
@inproceedings{Sanabria_breast_18,
author = {Sergio Sanabria and Katharina Martini and Marga Rominger and Konstantin Dedes and D. Vorburger and Thomas Frauenfelder and Orcun Goksel},
title = {Breast cancer assessment with hand-held ultrasound based Speed of Sound: preliminary results},
booktitle = {European Conference on Radiology (ECR)},
year = {2018},
doi = {10.1594/ecr2018/C-1388}
}
 [2018] M.-L. Metzger, Gregor Freystätter, Katharina Martini, Thomas Frauenfelder, Orcun Goksel, Sergio J. Sanabria, and Marga B. Rominger:
"Sarcopenia assessment with handheld ultrasound: a novel technique based on speed of sound",
In European Conference on Radiology (ECR), Vienna, Austria, Feb 2018.
[2018] M.-L. Metzger, Gregor Freystätter, Katharina Martini, Thomas Frauenfelder, Orcun Goksel, Sergio J. Sanabria, and Marga B. Rominger:
"Sarcopenia assessment with handheld ultrasound: a novel technique based on speed of sound",
In European Conference on Radiology (ECR), Vienna, Austria, Feb 2018.
@inproceedings{Metzger_sarcopenia_18,
author = {M-L Metzger and Gregor Freyst\"{a}tter and Katharina Martini and Thomas Frauenfelder and Orcun Goksel and Sergio J Sanabria and Marga B. Rominger},
title = {Sarcopenia assessment with handheld ultrasound: a novel technique based on speed of sound},
booktitle = {European Conference on Radiology (ECR)},
year = {2018},
doi = {10.1594/ecr2018/C-1211}
}
 [2018] Katharina Martini, Sergio J. Sanabria, Serafino Forte, Rahel A. Kubik-Huch, Thomas Frauenfelder, Orcun Goksel, and Marga B. Rominger:
"Hand-held speed-of-sound ultrasound -- a novel cost-efficient biomarker for breast density assessment",
In European Conference on Radiology (ECR), Vienna, Austria, Feb 2018.
[2018] Katharina Martini, Sergio J. Sanabria, Serafino Forte, Rahel A. Kubik-Huch, Thomas Frauenfelder, Orcun Goksel, and Marga B. Rominger:
"Hand-held speed-of-sound ultrasound -- a novel cost-efficient biomarker for breast density assessment",
In European Conference on Radiology (ECR), Vienna, Austria, Feb 2018.
@inproceedings{Martini_hand-held_18,
author = {Katharina Martini and Sergio J Sanabria and Serafino Forte and Rahel A Kubik-Huch and Thomas Frauenfelder and Orcun Goksel and Marga B. Rominger},
title = {Hand-held speed-of-sound ultrasound -- a novel cost-efficient biomarker for breast density assessment},
booktitle = {European Conference on Radiology (ECR)},
year = {2018},
doi = {10.1594/ecr2018/C-0037}
}
2017
 [2017] Sergio J. Sanabria, Marga B. Rominger, Corin F. Otesteanu, Farrukh I. Sheikh, Volker Klingmueller, and Orcun Goksel:
"Can speed of sound be better than conventional elastography for breast characterization?",
In Annual Meeting of Radiological Society of North America (RSNA), Chicago, IL, USA, Nov 2017.
[2017] Sergio J. Sanabria, Marga B. Rominger, Corin F. Otesteanu, Farrukh I. Sheikh, Volker Klingmueller, and Orcun Goksel:
"Can speed of sound be better than conventional elastography for breast characterization?",
In Annual Meeting of Radiological Society of North America (RSNA), Chicago, IL, USA, Nov 2017.
@inproceedings{Sanabria_can_17,
author = {Sergio J Sanabria and Marga B Rominger and Corin F Otesteanu and Farrukh I Sheikh and Volker Klingmueller and Orcun Goksel},
title = {Can speed of sound be better than conventional elastography for breast characterization?},
booktitle = {Annual Meeting of Radiological Society of North America (RSNA)},
year = {2017}
}
 [2017] Waleed Farrukh, Antonio Foncubierta-Rodriguez, Anca-Nicoleta Ciubotaru, Guillaume Jaume, Costas Bekas, Orcun Goksel, and Maria Gabrani:
"Interpreting data from scanned tables",
In GREC 2017: 12th IAPR International Workshop on Graphics Recognition, Kyoto, Japan, Nov 2017.
[2017] Waleed Farrukh, Antonio Foncubierta-Rodriguez, Anca-Nicoleta Ciubotaru, Guillaume Jaume, Costas Bekas, Orcun Goksel, and Maria Gabrani:
"Interpreting data from scanned tables",
In GREC 2017: 12th IAPR International Workshop on Graphics Recognition, Kyoto, Japan, Nov 2017.
@inproceedings{Farrukh_interpreting_17,
author = {Waleed Farrukh and Antonio Foncubierta-Rodriguez and Anca-Nicoleta Ciubotaru and Guillaume Jaume and Costas Bekas and Orcun Goksel and Maria Gabrani},
title = {Interpreting data from scanned tables},
booktitle = {GREC 2017: 12th IAPR International Workshop on Graphics Recognition},
year = {2017},
doi = {10.1109/ICDAR.2017.250}
}
 [2017] Pushpak Pati, Murat Arar, Govind Kaigala, Kashyap Aditya, Orcun Goksel, and Maria Gabrani:
"Computational Immunohistochemistry: Recipes for Standardization of Immunostaining",
In MICCAI, Quebec, QC, Canada, Sep 2017.
[2017] Pushpak Pati, Murat Arar, Govind Kaigala, Kashyap Aditya, Orcun Goksel, and Maria Gabrani:
"Computational Immunohistochemistry: Recipes for Standardization of Immunostaining",
In MICCAI, Quebec, QC, Canada, Sep 2017.
@inproceedings{Pati_computational_17,
author = {Pushpak Pati and Murat Arar and Govind Kaigala and Kashyap Aditya and Orcun Goksel and Maria Gabrani},
title = {Computational Immunohistochemistry: Recipes for Standardization of Immunostaining},
booktitle = {MICCAI},
year = {2017},
doi = {10.1007/978-3-319-66185-8_6}
}
 [2017] Corin F. Otesteanu, Bhaskara R. Chintada, Edoardo Mazza, Sergio J. Sanabria, and Orcun Goksel:
"Quantification of nonlinear elastic constants using polynomials in quasi-incompressible soft solids",
In IEEE Int Ultrasonics Symposium (IUS), Washington, DC, USA, Sep 2017.
[2017] Corin F. Otesteanu, Bhaskara R. Chintada, Edoardo Mazza, Sergio J. Sanabria, and Orcun Goksel:
"Quantification of nonlinear elastic constants using polynomials in quasi-incompressible soft solids",
In IEEE Int Ultrasonics Symposium (IUS), Washington, DC, USA, Sep 2017.
@inproceedings{Otesteanu_quantification_17,
author = {Corin F. Otesteanu and Bhaskara R. Chintada and Edoardo Mazza and Sergio J. Sanabria and Orcun Goksel},
title = {Quantification of nonlinear elastic constants using polynomials in quasi-incompressible soft solids},
booktitle = {IEEE Int Ultrasonics Symposium (IUS)},
year = {2017},
doi = {10.1109/ULTSYM.2017.8091737}
}
 [2017] Bhaskara R. Chintada, Sergio J. Sanabria, Wolfgang Bost, and Orcun Goksel:
"Reflector-Based 3D Tomographic Ultrasound Reconstruction: Simulation Study",
In IEEE Int Ultrasonics Symposium (IUS), Washington, DC, USA, Sep 2017.
[2017] Bhaskara R. Chintada, Sergio J. Sanabria, Wolfgang Bost, and Orcun Goksel:
"Reflector-Based 3D Tomographic Ultrasound Reconstruction: Simulation Study",
In IEEE Int Ultrasonics Symposium (IUS), Washington, DC, USA, Sep 2017.
@inproceedings{Chintada_reflector-based_17,
author = {Bhaskara R. Chintada and Sergio J. Sanabria and Wolfgang Bost and Orcun Goksel},
title = {Reflector-Based 3D Tomographic Ultrasound Reconstruction: Simulation Study},
booktitle = {IEEE Int Ultrasonics Symposium (IUS)},
year = {2017},
doi = {10.1109/ULTSYM.2017.8092257}
}
 [2017] Fabien Pean, Fabio Carrillo, Philipp Fuernstahl, and Orcun Goksel:
"Physical Simulation of the Interosseous Ligaments During Forearm Rotation",
In Computer Assisted Orthopaedic Surgery, Aachen, Germany, Jun 2017.
[2017] Fabien Pean, Fabio Carrillo, Philipp Fuernstahl, and Orcun Goksel:
"Physical Simulation of the Interosseous Ligaments During Forearm Rotation",
In Computer Assisted Orthopaedic Surgery, Aachen, Germany, Jun 2017.
@inproceedings{Pean_physical_17,
author = {Fabien Pean and Fabio Carrillo and Philipp Fuernstahl and Orcun Goksel},
title = {Physical Simulation of the Interosseous Ligaments During Forearm Rotation},
booktitle = {Computer Assisted Orthopaedic Surgery},
year = {2017},
url = {https://easychair.org/publications/paper/350665},
doi = {10.29007/74wg}
}
 [2017] A. Gomariz, P. Helbling, S. Isringhausen, U. Suessbier, A. Becker, A. Boss, T. Nagasawa, G. Paul, O. Goksel, G. Szekely, and others:
"Multiscale Image-Based Quantitative Analysis of Bone Marrow Stromal Network Topology Reveals Strict Spatial Constraints for Hematopoietic-Stromal Cellular Interactions",
In Haematologica, pp. 78-79, 2017.
[2017] A. Gomariz, P. Helbling, S. Isringhausen, U. Suessbier, A. Becker, A. Boss, T. Nagasawa, G. Paul, O. Goksel, G. Szekely, and others:
"Multiscale Image-Based Quantitative Analysis of Bone Marrow Stromal Network Topology Reveals Strict Spatial Constraints for Hematopoietic-Stromal Cellular Interactions",
In Haematologica, pp. 78-79, 2017.
@inproceedings{Gomariz_multiscale_17,
author = {Gomariz, A and Helbling, P and Isringhausen, S and Suessbier, U and Becker, A and Boss, A and Nagasawa, T and Paul, G and Goksel, O and Szekely, G and others},
title = {Multiscale Image-Based Quantitative Analysis of Bone Marrow Stromal Network Topology Reveals Strict Spatial Constraints for Hematopoietic-Stromal Cellular Interactions},
booktitle = {Haematologica},
year = {2017},
volume = {102},
pages = {78-79}
}
 [2017] Oliver Mattausch, Elisabeth Ren, Michael Bajka, Kenneth Vanhoey, and Orcun Goksel:
"Comparison of Texture Synthesis Methods for Content Generation in Ultrasound Training Simulation",
In SPIE Medical Imaging, pp. 1-8, Orlando, FL, USA, Feb 2017.
[2017] Oliver Mattausch, Elisabeth Ren, Michael Bajka, Kenneth Vanhoey, and Orcun Goksel:
"Comparison of Texture Synthesis Methods for Content Generation in Ultrasound Training Simulation",
In SPIE Medical Imaging, pp. 1-8, Orlando, FL, USA, Feb 2017.
@inproceedings{Mattausch_comparison_17,
author = {Oliver Mattausch and Elisabeth Ren and Michael Bajka and Kenneth Vanhoey and Orcun Goksel},
title = {Comparison of Texture Synthesis Methods for Content Generation in Ultrasound Training Simulation},
booktitle = {SPIE Medical Imaging},
year = {2017},
pages = {1-8},
doi = {10.1117/12.2250604}
}
2016
 [2016] Orcun Goksel, Valeria DeLuca, Maxim Makhinya, and Christine Tanner:
"Abdominal Motion Tracking and Prediction in 2D Ultrasound",
In 4D Treatment Planning (4DTP) Workshop, Groningen, Netherlands, Nov 2016.
[2016] Orcun Goksel, Valeria DeLuca, Maxim Makhinya, and Christine Tanner:
"Abdominal Motion Tracking and Prediction in 2D Ultrasound",
In 4D Treatment Planning (4DTP) Workshop, Groningen, Netherlands, Nov 2016.
@inproceedings{Goksel_abdominal_16,
author = {Orcun Goksel and Valeria DeLuca and Maxim Makhinya and Christine Tanner},
title = {Abdominal Motion Tracking and Prediction in 2D Ultrasound},
booktitle = {4D Treatment Planning (4DTP) Workshop},
year = {2016}
}
 [2016] Valeriy Vishnevskiy, Christine Tanner, and Orcun Goksel:
"Accurate and Fast Deformable Image Registration Allowing For Sliding Interfaces",
In 4D Treatment Planning (4DTP) Workshop, Groningen, Netherlands, Nov 2016.
[2016] Valeriy Vishnevskiy, Christine Tanner, and Orcun Goksel:
"Accurate and Fast Deformable Image Registration Allowing For Sliding Interfaces",
In 4D Treatment Planning (4DTP) Workshop, Groningen, Netherlands, Nov 2016.
@inproceedings{Vishnevskiy_accurate_16,
author = {Valeriy Vishnevskiy and Christine Tanner and Orcun Goksel},
title = {Accurate and Fast Deformable Image Registration Allowing For Sliding Interfaces},
booktitle = {4D Treatment Planning (4DTP) Workshop},
year = {2016}
}
 [2016] Sergio J. Sanabria and Orcun Goksel:
"Hand-held Sound-Speed Mammography Based on Ultrasound Reflector Tracking",
In MICCAI, pp. 568-576, Athens, Greece, Oct 2016.
[2016] Sergio J. Sanabria and Orcun Goksel:
"Hand-held Sound-Speed Mammography Based on Ultrasound Reflector Tracking",
In MICCAI, pp. 568-576, Athens, Greece, Oct 2016.
@inproceedings{Sanabria_hand-held_16,
author = {Sergio J. Sanabria and Orcun Goksel},
title = {Hand-held Sound-Speed Mammography Based on Ultrasound Reflector Tracking},
booktitle = {MICCAI},
year = {2016},
pages = {568-576},
doi = {10.1007/978-3-319-46720-7_66}
}
 [2016] Firat Ozdemir, Ece Ozkan, and Orcun Goksel:
"Graphical Modeling of Ultrasound Propagation in Tissue for Automatic Bone Segmentation",
In MICCAI, pp. 256-264, Athens, Greece, Oct 2016.
[2016] Firat Ozdemir, Ece Ozkan, and Orcun Goksel:
"Graphical Modeling of Ultrasound Propagation in Tissue for Automatic Bone Segmentation",
In MICCAI, pp. 256-264, Athens, Greece, Oct 2016.
@inproceedings{Ozdemir_graphical_16,
author = {Firat Ozdemir and Ece Ozkan and Orcun Goksel},
title = {Graphical Modeling of Ultrasound Propagation in Tissue for Automatic Bone Segmentation},
booktitle = {MICCAI},
year = {2016},
pages = {256-264},
doi = {10.1007/978-3-319-46723-8_30}
}
 [2016] Christine Tanner, Celine Eggenberger, Barbara Flach, Oliver Mattausch, Michael Bajka, and Orcun Goksel:
"4D Reconstruction of Fetal Heart Ultrasound Images in Presence of Fetal Motion",
In MICCAI, pp. 593-601, Athens, Greece, Oct 2016.
[2016] Christine Tanner, Celine Eggenberger, Barbara Flach, Oliver Mattausch, Michael Bajka, and Orcun Goksel:
"4D Reconstruction of Fetal Heart Ultrasound Images in Presence of Fetal Motion",
In MICCAI, pp. 593-601, Athens, Greece, Oct 2016.
@inproceedings{Tanner_4D_16,
author = {Christine Tanner and Celine Eggenberger and Barbara Flach and Oliver Mattausch and Michael Bajka and Orcun Goksel},
title = {4D Reconstruction of Fetal Heart Ultrasound Images in Presence of Fetal Motion},
booktitle = {MICCAI},
year = {2016},
pages = {593-601},
doi = {10.1007/978-3-319-46720-7_69}
}
 [2016] Oliver Mattausch and Orcun Goksel:
"Image-based PSF Estimation for Ultrasound Training Simulation",
In MICCAI W Simulation and Synthesis in Medical Imaging (SASHIMI), pp. 23-33, Athens, Greece, Oct 2016.
[2016] Oliver Mattausch and Orcun Goksel:
"Image-based PSF Estimation for Ultrasound Training Simulation",
In MICCAI W Simulation and Synthesis in Medical Imaging (SASHIMI), pp. 23-33, Athens, Greece, Oct 2016.
@inproceedings{Mattausch_image-based_16,
author = {Oliver Mattausch and Orcun Goksel},
title = {Image-based PSF Estimation for Ultrasound Training Simulation},
booktitle = {MICCAI W Simulation and Synthesis in Medical Imaging (SASHIMI)},
year = {2016},
pages = {23-33},
doi = {10.1007/978-3-319-46630-9_3}
}
 [2016] Barbara Flach, Maxim Makhinya, and Orcun Goksel:
"PURE: Panoramic Ultrasound Reconstruction by Seamless Stitching of Volumes",
In MICCAI W Simulation and Synthesis in Medical Imaging (SASHIMI), pp. 75-84, Athens, Greece, Oct 2016.
[2016] Barbara Flach, Maxim Makhinya, and Orcun Goksel:
"PURE: Panoramic Ultrasound Reconstruction by Seamless Stitching of Volumes",
In MICCAI W Simulation and Synthesis in Medical Imaging (SASHIMI), pp. 75-84, Athens, Greece, Oct 2016.
@inproceedings{Flach_pure_16,
author = {Barbara Flach and Maxim Makhinya and Orcun Goksel},
title = {PURE: Panoramic Ultrasound Reconstruction by Seamless Stitching of Volumes},
booktitle = {MICCAI W Simulation and Synthesis in Medical Imaging (SASHIMI)},
year = {2016},
pages = {75-84},
doi = {10.1007/978-3-319-46630-9_8}
}
 [2016] Janine Thoma, Firat Ozdemir, and Orcun Goksel:
"Automatic Segmentation of Abdominal MRI Using Selective Sampling and Random Walker",
In MICCAI W Medical Computer Vision (MCV), pp. 83-93, Athens, Greece, Oct 2016.
[2016] Janine Thoma, Firat Ozdemir, and Orcun Goksel:
"Automatic Segmentation of Abdominal MRI Using Selective Sampling and Random Walker",
In MICCAI W Medical Computer Vision (MCV), pp. 83-93, Athens, Greece, Oct 2016.
@inproceedings{Thoma_automatic_16,
author = {Janine Thoma and Firat Ozdemir and Orcun Goksel},
title = {Automatic Segmentation of Abdominal MRI Using Selective Sampling and Random Walker},
booktitle = {MICCAI W Medical Computer Vision (MCV)},
year = {2016},
pages = {83-93},
doi = {10.1007/978-3-319-61188-4_8}
}
 [2016] Sergio J. Sanabria, Marga Rominger, Corin F. Otesteanu, Edoardo Mazza, and Orcun Goksel:
"Non-Linear Characterization of the Liver by Combining Shear and Longitudinal Wave Speed With Strain Observations",
In Int Tissue Elasticity Conf (ITEC), Vermont, NE, USA, Oct 2016.
[2016] Sergio J. Sanabria, Marga Rominger, Corin F. Otesteanu, Edoardo Mazza, and Orcun Goksel:
"Non-Linear Characterization of the Liver by Combining Shear and Longitudinal Wave Speed With Strain Observations",
In Int Tissue Elasticity Conf (ITEC), Vermont, NE, USA, Oct 2016.
@inproceedings{Sanabria_non-linear_16,
author = {Sergio J Sanabria and Marga Rominger and Corin F. Otesteanu and Edoardo Mazza and Orcun Goksel},
title = {Non-Linear Characterization of the Liver by Combining Shear and Longitudinal Wave Speed With Strain Observations},
booktitle = {Int Tissue Elasticity Conf (ITEC)},
year = {2016}
}
 [2016] Ece Ozkan and Orcun Goksel:
"Compliance Boundary Conditions for Elasticity Reconstruction Using FEM Inverse Problem",
In Int Tissue Elasticity Conf (ITEC), Vermont, NE, USA, Oct 2016.
[2016] Ece Ozkan and Orcun Goksel:
"Compliance Boundary Conditions for Elasticity Reconstruction Using FEM Inverse Problem",
In Int Tissue Elasticity Conf (ITEC), Vermont, NE, USA, Oct 2016.
@inproceedings{Ozkan_compliance_16,
author = {Ece Ozkan and Orcun Goksel},
title = {Compliance Boundary Conditions for Elasticity Reconstruction Using FEM Inverse Problem},
booktitle = {Int Tissue Elasticity Conf (ITEC)},
year = {2016}
}
 [2016] Corin F. Otesteanu, Sergio J. Sanabria, and Orcun Goksel:
"Low Cost Alternative for Ultrasound Harmonic Elastography",
In Int Tissue Elasticity Conf (ITEC), Vermont, NE, USA, Oct 2016.
[2016] Corin F. Otesteanu, Sergio J. Sanabria, and Orcun Goksel:
"Low Cost Alternative for Ultrasound Harmonic Elastography",
In Int Tissue Elasticity Conf (ITEC), Vermont, NE, USA, Oct 2016.
@inproceedings{Otesteanu_low-cost_16,
author = {Corin F. Otesteanu and Sergio J Sanabria and Orcun Goksel},
title = {Low Cost Alternative for Ultrasound Harmonic Elastography},
booktitle = {Int Tissue Elasticity Conf (ITEC)},
year = {2016}
}
 [2016] Andreas Kurth, Andreas Tretter, Pascal A. Hager, Sergio Sanabria, Orcun Goksel, Lothar Thiele, and Luca Benini:
"Mobile Ultrasound Imaging on Heterogeneous Multi-Core Platform",
In ACM/IEEE Symp on Embedded Systems for Real-time Multimedia (ESTIMedia), pp. 9-18, Pittsburgh, USA, Oct 2016.
[2016] Andreas Kurth, Andreas Tretter, Pascal A. Hager, Sergio Sanabria, Orcun Goksel, Lothar Thiele, and Luca Benini:
"Mobile Ultrasound Imaging on Heterogeneous Multi-Core Platform",
In ACM/IEEE Symp on Embedded Systems for Real-time Multimedia (ESTIMedia), pp. 9-18, Pittsburgh, USA, Oct 2016.
* Best Paper Award
@inproceedings{Kurth_mobile_16,
author = {Andreas Kurth and Andreas Tretter and Pascal A. Hager and Sergio Sanabria and Orcun Goksel and Lothar Thiele and Luca Benini},
title = {Mobile Ultrasound Imaging on Heterogeneous Multi-Core Platform},
booktitle = {ACM/IEEE Symp on Embedded Systems for Real-time Multimedia (ESTIMedia)},
year = {2016},
pages = {9-18},
doi = {10.1145/2993452.2993565}
}
 [2016] Ece Ozkan and Orcun Goksel:
"Inverse Problem of Ultrasound Beamforming with Sparsity in Time and Frequency Domain",
In IEEE Int Ultrasonics Symp (IUS), Tours, France, Sep 2016.
[2016] Ece Ozkan and Orcun Goksel:
"Inverse Problem of Ultrasound Beamforming with Sparsity in Time and Frequency Domain",
In IEEE Int Ultrasonics Symp (IUS), Tours, France, Sep 2016.
@inproceedings{Ozkan_inverse_16,
author = {Ece Ozkan and Orcun Goksel},
title = {Inverse Problem of Ultrasound Beamforming with Sparsity in Time and Frequency Domain},
booktitle = {IEEE Int Ultrasonics Symp (IUS)},
year = {2016},
doi = {10.1109/ULTSYM.2016.7728892}
}
 [2016] Oliver Mattausch and Orcun Goksel:
"Monte-Carlo Ray-Tracing for Realistic Interactive Ultrasound Simulation",
In Eurographics W Visual Computing for Biology and Medicine (VCBM), pp. 1-9, Bergen, Norway, Sep 2016.
[2016] Oliver Mattausch and Orcun Goksel:
"Monte-Carlo Ray-Tracing for Realistic Interactive Ultrasound Simulation",
In Eurographics W Visual Computing for Biology and Medicine (VCBM), pp. 1-9, Bergen, Norway, Sep 2016.
@inproceedings{Mattausch_monte-carlo_16,
author = {Oliver Mattausch and Orcun Goksel},
title = {Monte-Carlo Ray-Tracing for Realistic Interactive Ultrasound Simulation},
booktitle = {Eurographics W Visual Computing for Biology and Medicine (VCBM)},
year = {2016},
pages = {1-9},
doi = {dx.doi.org/10.2312/vcbm.20161285}
}
 [2016] Xiaoran Chen, Christine Tanner, Orccun Göksel, Gábor Székely, and Valeria Luca:
"Temporal Prediction of Respiratory Motion Using a Trained Ensemble of Forecasting Methods",
In Medical Imaging and Augmented Reality (MIAR), pp. 383-391, Bern, Switzerland, Aug 2016.
[2016] Xiaoran Chen, Christine Tanner, Orccun Göksel, Gábor Székely, and Valeria Luca:
"Temporal Prediction of Respiratory Motion Using a Trained Ensemble of Forecasting Methods",
In Medical Imaging and Augmented Reality (MIAR), pp. 383-391, Bern, Switzerland, Aug 2016.
@inproceedings{Chen_temporal_16,
author = {Chen, Xiaoran and Tanner, Christine and G\"oksel, Or\ccun and Sz\'ekely, G\'abor and Luca, Valeria},
title = {Temporal Prediction of Respiratory Motion Using a Trained Ensemble of Forecasting Methods},
booktitle = {Medical Imaging and Augmented Reality (MIAR)},
year = {2016},
pages = {383-391},
doi = {10.1007/978-3-319-43775-0_35}
}
 [2016] Corin Felix Otesteanu, Sergio J. Sanabria, and Orcun Goksel:
"Analysis of Excitation Frequency in Elasticity Reconstruction Using the FEM Inverse-Problem",
In IEEE Int Symp Biomedical Imaging (ISBI), pp. 485-8, Prague, Czech Republic, Apr 2016.
[2016] Corin Felix Otesteanu, Sergio J. Sanabria, and Orcun Goksel:
"Analysis of Excitation Frequency in Elasticity Reconstruction Using the FEM Inverse-Problem",
In IEEE Int Symp Biomedical Imaging (ISBI), pp. 485-8, Prague, Czech Republic, Apr 2016.
@inproceedings{Otesteanu_analysis_16,
author = {Corin Felix Otesteanu and Sergio J Sanabria and Orcun Goksel},
title = {Analysis of Excitation Frequency in Elasticity Reconstruction Using the FEM Inverse-Problem},
booktitle = {IEEE Int Symp Biomedical Imaging (ISBI)},
year = {2016},
pages = {485-8},
doi = {10.1109/ISBI.2016.7493313}
}
 [2016] Orcun Goksel, Valery Vishnevsky, Alvaro Gomariz Carrillo, and Christine Tanner:
"Imaging of Sliding Visceral Interfaces During Breathing",
In IEEE Int Symp Biomedical Imaging (ISBI), pp. 298-301, Prague, Czech Republic, Apr 2016.
[2016] Orcun Goksel, Valery Vishnevsky, Alvaro Gomariz Carrillo, and Christine Tanner:
"Imaging of Sliding Visceral Interfaces During Breathing",
In IEEE Int Symp Biomedical Imaging (ISBI), pp. 298-301, Prague, Czech Republic, Apr 2016.
@inproceedings{Goksel_imaging_16,
author = {Orcun Goksel and Valery Vishnevsky and Alvaro Gomariz Carrillo and Christine Tanner},
title = {Imaging of Sliding Visceral Interfaces During Breathing},
booktitle = {IEEE Int Symp Biomedical Imaging (ISBI)},
year = {2016},
pages = {298-301},
doi = {10.1109/ISBI.2016.7493268}
}
 [2016] Barbara Flach, Maxim Makhinya, and Orcun Goksel:
"Model-based Compensation of Tissue Deformation During Data Acquisition for Interpolative Ultrasound Simulation",
In IEEE Int Symp Biomed Imaging (ISBI), pp. 502-5, Prague, Czech Republic, Apr 2016.
[2016] Barbara Flach, Maxim Makhinya, and Orcun Goksel:
"Model-based Compensation of Tissue Deformation During Data Acquisition for Interpolative Ultrasound Simulation",
In IEEE Int Symp Biomed Imaging (ISBI), pp. 502-5, Prague, Czech Republic, Apr 2016.
@inproceedings{Flach_model-based_16,
author = {Barbara Flach and Maxim Makhinya and Orcun Goksel},
title = {Model-based Compensation of Tissue Deformation During Data Acquisition for Interpolative Ultrasound Simulation},
booktitle = {IEEE Int Symp Biomed Imaging (ISBI)},
year = {2016},
pages = {502-5},
doi = {10.1109/ISBI.2016.7493317}
}
2015 and Earlier
 [2015] Maxim Makhinya and Orcun Goksel:
"Real-time Tracking of Liver Landmarks in 2D Ultrasound Sequences",
In 4D Treatment Planning (4DTP) Workshop, Dresden, Germany, Nov 2015.
[2015] Maxim Makhinya and Orcun Goksel:
"Real-time Tracking of Liver Landmarks in 2D Ultrasound Sequences",
In 4D Treatment Planning (4DTP) Workshop, Dresden, Germany, Nov 2015.
@inproceedings{Makhinya_real-time_15,
author = {Maxim Makhinya and Orcun Goksel},
title = {Real-time Tracking of Liver Landmarks in 2D Ultrasound Sequences},
booktitle = {4D Treatment Planning (4DTP) Workshop},
year = {2015}
}
 [2015] Maxim Makhinya and Orcun Goksel:
"Motion Tracking in 2D Ultrasound Using Vessel Models and Robust Optic-Flow",
In MICCAI Challenge on Liver Motion Tracking (CLUST), Munich, Germany, Oct 2015.
[2015] Maxim Makhinya and Orcun Goksel:
"Motion Tracking in 2D Ultrasound Using Vessel Models and Robust Optic-Flow",
In MICCAI Challenge on Liver Motion Tracking (CLUST), Munich, Germany, Oct 2015.
@inproceedings{Makhinya_motion_15,
author = {Maxim Makhinya and Orcun Goksel},
title = {Motion Tracking in 2D Ultrasound Using Vessel Models and Robust Optic-Flow},
booktitle = {MICCAI Challenge on Liver Motion Tracking (CLUST)},
year = {2015},
url = {http://clust.ethz.ch/opendownload/CLUST2015/makhinya_clust15.pdf}
}
 [2015] Sergio J. Sanabria, Corin F. Otesteanu, and Orcun Goksel:
"Hand-held Sound-speed Imaging for the Reconstruction of Bulk Modulus and Poisson Ratio in a Commercial Tissue-Mimicking Phantom",
In Int Tissue Elasticity Conf (ITEC), Verona, Italy, Sep 2015.
[2015] Sergio J. Sanabria, Corin F. Otesteanu, and Orcun Goksel:
"Hand-held Sound-speed Imaging for the Reconstruction of Bulk Modulus and Poisson Ratio in a Commercial Tissue-Mimicking Phantom",
In Int Tissue Elasticity Conf (ITEC), Verona, Italy, Sep 2015.
@inproceedings{Sanabria_hand-held_15,
author = {Sergio J. Sanabria and Corin F. Otesteanu and Orcun Goksel},
title = {Hand-held Sound-speed Imaging for the Reconstruction of Bulk Modulus and Poisson Ratio in a Commercial Tissue-Mimicking Phantom},
booktitle = {Int Tissue Elasticity Conf (ITEC)},
year = {2015}
}
 [2015] Sergio J. Sanabria and Orcun Goksel:
"Total-Variation Regularization of Hand-held Limited-Angle Sound-Speed Tomography Based on Coherent Reflector for Breast Cancer Detection",
In Int Tissue Elasticity Conf (ITEC), Verona, Italy, Sep 2015.
[2015] Sergio J. Sanabria and Orcun Goksel:
"Total-Variation Regularization of Hand-held Limited-Angle Sound-Speed Tomography Based on Coherent Reflector for Breast Cancer Detection",
In Int Tissue Elasticity Conf (ITEC), Verona, Italy, Sep 2015.
@inproceedings{Sanabria_total-variation_15,
author = {Sergio J. Sanabria and Orcun Goksel},
title = {Total-Variation Regularization of Hand-held Limited-Angle Sound-Speed Tomography Based on Coherent Reflector for Breast Cancer Detection},
booktitle = {Int Tissue Elasticity Conf (ITEC)},
year = {2015}
}
 [2015] Ece Ozkan and Orcun Goksel:
"Compliance Boundary Conditions for Simulating Deformations in a Limited Region of Interest",
In IEEE Eng Medicine and Biology Conf (EMBC), pp. 929-32, Milano, Italy, Aug 2015.
[2015] Ece Ozkan and Orcun Goksel:
"Compliance Boundary Conditions for Simulating Deformations in a Limited Region of Interest",
In IEEE Eng Medicine and Biology Conf (EMBC), pp. 929-32, Milano, Italy, Aug 2015.
@inproceedings{Ozkan_compliance_15,
author = {Ece Ozkan and Orcun Goksel},
title = {Compliance Boundary Conditions for Simulating Deformations in a Limited Region of Interest},
booktitle = {IEEE Eng Medicine and Biology Conf (EMBC)},
year = {2015},
pages = {929-32},
doi = {10.1109/EMBC.2015.7318515}
}
 [2015] Oliver Mattausch and Orcun Goksel:
"Scatterer Reconstruction and Parametrization of Homogeneous Tissue for Ultrasound Image Simulation",
In IEEE Eng Medicine and Biology Conf (EMBC), pp. 6350-3, Milano, Italy, Aug 2015.
[2015] Oliver Mattausch and Orcun Goksel:
"Scatterer Reconstruction and Parametrization of Homogeneous Tissue for Ultrasound Image Simulation",
In IEEE Eng Medicine and Biology Conf (EMBC), pp. 6350-3, Milano, Italy, Aug 2015.
@inproceedings{Mattausch_scatterer_15,
author = {Oliver Mattausch and Orcun Goksel},
title = {Scatterer Reconstruction and Parametrization of Homogeneous Tissue for Ultrasound Image Simulation},
booktitle = {IEEE Eng Medicine and Biology Conf (EMBC)},
year = {2015},
pages = {6350-3},
doi = {10.1109/EMBC.2015.7319845}
}
 [2015] Peter Baki, Sergio J. Sanabria, Gabor Kosa, Gabor Szekely, and Orcun Goksel:
"Thermal Expansion Imaging for Monitoring Lesion Depth using M-Mode Ultrasound during Cardiac RF Ablation: In-vitro Study",
In Information Processing in Computer-Assisted Interventions (IPCAI), Barcelona, Spain, Jun 2015.
[2015] Peter Baki, Sergio J. Sanabria, Gabor Kosa, Gabor Szekely, and Orcun Goksel:
"Thermal Expansion Imaging for Monitoring Lesion Depth using M-Mode Ultrasound during Cardiac RF Ablation: In-vitro Study",
In Information Processing in Computer-Assisted Interventions (IPCAI), Barcelona, Spain, Jun 2015.
@inproceedings{Baki_thermal_15t,
author = {Peter Baki and Sergio J. Sanabria and Gabor Kosa and Gabor Szekely and Orcun Goksel},
title = {Thermal Expansion Imaging for Monitoring Lesion Depth using M-Mode Ultrasound during Cardiac RF Ablation: In-vitro Study},
booktitle = {Information Processing in Computer-Assisted Interventions (IPCAI)},
year = {2015}
}
 [2015] Valery Vishnevsky, Tobias Gass, Gabor Szekely, Christine Tanner, and Orcun Goksel:
"Unsupervised Detection of Local Errors in Image Registration",
In IEEE Int Symp Biomedical Imaging (ISBI), pp. 841-4, New York, USA, Apr 2015.
[2015] Valery Vishnevsky, Tobias Gass, Gabor Szekely, Christine Tanner, and Orcun Goksel:
"Unsupervised Detection of Local Errors in Image Registration",
In IEEE Int Symp Biomedical Imaging (ISBI), pp. 841-4, New York, USA, Apr 2015.
@inproceedings{Vishnevsky_unsupervised_15,
author = {Valery Vishnevsky and Tobias Gass and Gabor Szekely and Christine Tanner and Orcun Goksel},
title = {Unsupervised Detection of Local Errors in Image Registration},
booktitle = {IEEE Int Symp Biomedical Imaging (ISBI)},
year = {2015},
pages = {841-4},
doi = {10.1109/ISBI.2015.7164002}
}
 [2014] Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Multi-Atlas Segmentation and Landmark Localization in Images with Large Fields of View",
In MICCAI W Medical Computer Vision (MCV): Algorithms for Big Data, pp. 171-80, Boston, MA, USA, Sep 2014.
[2014] Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Multi-Atlas Segmentation and Landmark Localization in Images with Large Fields of View",
In MICCAI W Medical Computer Vision (MCV): Algorithms for Big Data, pp. 171-80, Boston, MA, USA, Sep 2014.
@inproceedings{Gass_multi-atlas_14,
author = {Tobias Gass and Gabor Szekely and Orcun Goksel},
title = {Multi-Atlas Segmentation and Landmark Localization in Images with Large Fields of View},
booktitle = {MICCAI W Medical Computer Vision (MCV): Algorithms for Big Data},
year = {2014},
pages = {171-80},
doi = {10.1007/978-3-319-13972-2_16}
}
 [2014] Valeriy Vishnevskiy, Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Total Variation Regularization of Displacements in Parametric Image Registration",
In MICCAI W Abdominal Imaging: Computational and Clinical Apps, pp. 211-220, Boston, MA, USA, Sep 2014.
[2014] Valeriy Vishnevskiy, Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Total Variation Regularization of Displacements in Parametric Image Registration",
In MICCAI W Abdominal Imaging: Computational and Clinical Apps, pp. 211-220, Boston, MA, USA, Sep 2014.
* Best Poster Award at the 4D Treatment Planning Workshop in London, UK (Nov, 2014)
@inproceedings{Vishnevskiy_total_14,
author = {Valeriy Vishnevskiy and Tobias Gass and Gabor Szekely and Orcun Goksel},
title = {Total Variation Regularization of Displacements in Parametric Image Registration},
booktitle = {MICCAI W Abdominal Imaging: Computational and Clinical Apps},
year = {2014},
pages = {211-220},
doi = {10.1007/978-3-319-13692-9_20}
}
 [2014] Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Registration Fusion using Markov Random Fields",
In Workshop on Biomedical Image Registration (WBIR), pp. 213-222, London, UK, Jul 2014.
[2014] Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Registration Fusion using Markov Random Fields",
In Workshop on Biomedical Image Registration (WBIR), pp. 213-222, London, UK, Jul 2014.
@inproceedings{Gass_registration_14,
author = {Tobias Gass and Gabor Szekely and Orcun Goksel},
title = {Registration Fusion using Markov Random Fields},
booktitle = {Workshop on Biomedical Image Registration (WBIR)},
year = {2014},
pages = {213-222},
doi = {10.1007/978-3-319-08554-8_22}
}
 [2014] Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Consistent Dense Correspondences from Pairwise Registrations",
In SHAPE Symp on Statistical Shape Models and Applications, pp. 10, Delemont, Switzerland, Jun 2014.
podium
[2014] Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Consistent Dense Correspondences from Pairwise Registrations",
In SHAPE Symp on Statistical Shape Models and Applications, pp. 10, Delemont, Switzerland, Jun 2014.
podium
@inproceedings{Gass_consistent_14,
author = {Tobias Gass and Gabor Szekely and Orcun Goksel},
title = {Consistent Dense Correspondences from Pairwise Registrations},
booktitle = {SHAPE Symp on Statistical Shape Models and Applications},
year = {2014},
pages = {10}
}
 [2014] Orcun Goksel, Tobias Gass, and Gabor Szekely:
"Segmentation and Landmark Localization Based on Multiple Atlases",
In IEEE ISBI VISCERAL Challenge, pp. 37-43, Beijing, China, May 2014.
[2014] Orcun Goksel, Tobias Gass, and Gabor Szekely:
"Segmentation and Landmark Localization Based on Multiple Atlases",
In IEEE ISBI VISCERAL Challenge, pp. 37-43, Beijing, China, May 2014.
@inproceedings{Goksel_segmentation_14,
author = {Orcun Goksel and Tobias Gass and Gabor Szekely},
title = {Segmentation and Landmark Localization Based on Multiple Atlases},
booktitle = {IEEE ISBI VISCERAL Challenge},
year = {2014},
number = {1194},
pages = {37-43},
url = {http://ceur-ws.org/Vol-1194/visceralISBI14-5.pdf}
}
 [2014] Alessandro Crimi, Maxim Makhinya, Ulrich Baumann, Gabor Szekely, and Orcun Goksel:
"Vessel tracking for Ultrasound-based venous pressure measurement",
In IEEE Int Symp Biomedical Imaging (ISBI), pp. 306-9, Beijing, China, Apr 2014.
[2014] Alessandro Crimi, Maxim Makhinya, Ulrich Baumann, Gabor Szekely, and Orcun Goksel:
"Vessel tracking for Ultrasound-based venous pressure measurement",
In IEEE Int Symp Biomedical Imaging (ISBI), pp. 306-9, Beijing, China, Apr 2014.
@inproceedings{Crimi_vessel_14,
author = {Alessandro Crimi and Maxim Makhinya and Ulrich Baumann and Gabor Szekely and Orcun Goksel},
title = {Vessel tracking for Ultrasound-based venous pressure measurement},
booktitle = {IEEE Int Symp Biomedical Imaging (ISBI)},
year = {2014},
pages = {306-9},
doi = {10.1109/ISBI.2014.6867870}
}
 [2014] Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Detection and correction of inconsistency-based errors in non-rigid registration",
In SPIE Medical Imaging, pp. 90341B, San Diego, CA, USA, Feb 2014.
podium
[2014] Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Detection and correction of inconsistency-based errors in non-rigid registration",
In SPIE Medical Imaging, pp. 90341B, San Diego, CA, USA, Feb 2014.
podium
@inproceedings{Gass_detection_14,
author = {Tobias Gass and Gabor Szekely and Orcun Goksel},
title = {Detection and correction of inconsistency-based errors in non-rigid registration},
booktitle = {SPIE Medical Imaging},
year = {2014},
pages = {90341B},
doi = {10.1117/12.2042757}
}
 [2014] Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Auxiliary anatomical labels for joint segmentation and atlas registration",
In SPIE Medical Imaging, pp. 90343T, San Diego, CA, USA, Feb 2014.
poster
[2014] Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Auxiliary anatomical labels for joint segmentation and atlas registration",
In SPIE Medical Imaging, pp. 90343T, San Diego, CA, USA, Feb 2014.
poster
@inproceedings{Gass_auxiliary_14,
author = {Tobias Gass and Gabor Szekely and Orcun Goksel},
title = {Auxiliary anatomical labels for joint segmentation and atlas registration},
booktitle = {SPIE Medical Imaging},
year = {2014},
pages = {90343T},
doi = {10.1117/12.2042876}
}
 [2013] Orçun Göksel and Gabor Székely:
"Improving FEM Inverse Problem Reconstructions By Incorporating All Displacement Observations Using Element Shape Function Interpolations",
In Int Tissue Elasticity Conf (ITEC), pp. 78, Lingfield, UK, Oct 2013.
podium
[2013] Orçun Göksel and Gabor Székely:
"Improving FEM Inverse Problem Reconstructions By Incorporating All Displacement Observations Using Element Shape Function Interpolations",
In Int Tissue Elasticity Conf (ITEC), pp. 78, Lingfield, UK, Oct 2013.
podium
@inproceedings{Goksel_improving_13,
author = {Or\c{c}un G\"oksel and Gabor Sz\'ekely},
title = {Improving FEM Inverse Problem Reconstructions By Incorporating All Displacement Observations Using Element Shape Function Interpolations},
booktitle = {Int Tissue Elasticity Conf (ITEC)},
year = {2013},
pages = {78},
url = {http://www.elasticityconference.org/PDF/2013/2013ITECProceedings.pdf}
}
 [2013] Péter Baki, Gabor Székely, and Orçun Göksel:
"Thermal Expansion Imaging For Real-time Lesion Depth Assessment During RF Catheter Ablation",
In Int Tissue Elasticity Conf (ITEC), pp. 70, Lingfield, UK, Oct 2013.
podium
[2013] Péter Baki, Gabor Székely, and Orçun Göksel:
"Thermal Expansion Imaging For Real-time Lesion Depth Assessment During RF Catheter Ablation",
In Int Tissue Elasticity Conf (ITEC), pp. 70, Lingfield, UK, Oct 2013.
podium
@inproceedings{Baki_thermal_13,
author = {P\'eter Baki and Gabor Sz\'ekely and Or\c{c}un G\"oksel},
title = {Thermal Expansion Imaging For Real-time Lesion Depth Assessment During RF Catheter Ablation},
booktitle = {Int Tissue Elasticity Conf (ITEC)},
year = {2013},
pages = {70},
url = {http://www.elasticityconference.org/PDF/2013/2013ITECProceedings.pdf}
}
 [2013] Kamal Shahim, Orcun Goksel, Philipp Jürgens, and Mauricio Reyes:
"Accuracy Improvement In Cranio-Maxillofacial Soft Tissue Simulation Using A Muscle Embedded Meshing Approach",
In IEEE Eng Medicine and Biology Conf (EMBC), pp. 7156-9, Osaka, Japan, Jul 2013.
[2013] Kamal Shahim, Orcun Goksel, Philipp Jürgens, and Mauricio Reyes:
"Accuracy Improvement In Cranio-Maxillofacial Soft Tissue Simulation Using A Muscle Embedded Meshing Approach",
In IEEE Eng Medicine and Biology Conf (EMBC), pp. 7156-9, Osaka, Japan, Jul 2013.
@inproceedings{Shahim_accuracy_13,
author = {Kamal Shahim and Orcun Goksel and Philipp J\"urgens and Mauricio Reyes},
title = {Accuracy Improvement In Cranio-Maxillofacial Soft Tissue Simulation Using A Muscle Embedded Meshing Approach},
booktitle = {IEEE Eng Medicine and Biology Conf (EMBC)},
year = {2013},
pages = {7156-9},
doi = {10.1109/EMBC.2013.6611208}
}
 [2013] Hua Ma, Thomas Coradi, Gabor Szekely, Benjamin Haas, and Orcun Goksel:
"Supervised Learning with Global Features for Image Retrieval in Atlas-Based Segmentation of Thoracic CT",
In Int Congress on Computer Assisted Radiology and Surgery (CARS), pp. S302, Heidelberg, Germany, Jun 2013.
poster
[2013] Hua Ma, Thomas Coradi, Gabor Szekely, Benjamin Haas, and Orcun Goksel:
"Supervised Learning with Global Features for Image Retrieval in Atlas-Based Segmentation of Thoracic CT",
In Int Congress on Computer Assisted Radiology and Surgery (CARS), pp. S302, Heidelberg, Germany, Jun 2013.
poster
@inproceedings{Ma_supervised_13,
author = {Hua Ma and Thomas Coradi and Gabor Szekely and Benjamin Haas and Orcun Goksel},
title = {Supervised Learning with Global Features for Image Retrieval in Atlas-Based Segmentation of Thoracic CT},
booktitle = {Int Congress on Computer Assisted Radiology and Surgery (CARS)},
year = {2013},
pages = {S302}
}
 [2013] Orcun Goksel, Tobias Gass, Valery Vishnevsky, and Gabor Szekely:
"Estimation of Atlas-Based Segmentation Outcome: Leveraging Information From Unsegmented Images",
In IEEE Int Symp Biomedical Imaging (ISBI), pp. 1203-6, San Francisco, CA, USA, Apr 2013.
podium
[2013] Orcun Goksel, Tobias Gass, Valery Vishnevsky, and Gabor Szekely:
"Estimation of Atlas-Based Segmentation Outcome: Leveraging Information From Unsegmented Images",
In IEEE Int Symp Biomedical Imaging (ISBI), pp. 1203-6, San Francisco, CA, USA, Apr 2013.
podium
@inproceedings{Goksel_estimation_13,
author = {Orcun Goksel and Tobias Gass and Valery Vishnevsky and Gabor Szekely},
title = {Estimation of Atlas-Based Segmentation Outcome: Leveraging Information From Unsegmented Images},
booktitle = {IEEE Int Symp Biomedical Imaging (ISBI)},
year = {2013},
pages = {1203-6},
doi = {10.1109/ISBI.2013.6556699}
}
 [2013] Orcun Goksel, Seokhee Jeon, Matthias Harders, and Gabor Szekely:
"Deformable Haptic Model Generation Through Manual Exploration",
In IEEE World Haptics Conference, pp. 543-8, Daejeon, South Korea, Apr 2013.
podium
[2013] Orcun Goksel, Seokhee Jeon, Matthias Harders, and Gabor Szekely:
"Deformable Haptic Model Generation Through Manual Exploration",
In IEEE World Haptics Conference, pp. 543-8, Daejeon, South Korea, Apr 2013.
podium
@inproceedings{Goksel_deformable_13,
author = {Orcun Goksel and Seokhee Jeon and Matthias Harders and Gabor Szekely},
title = {Deformable Haptic Model Generation Through Manual Exploration},
booktitle = {IEEE World Haptics Conference},
year = {2013},
pages = {543-8},
doi = {10.1109/WHC.2013.6548466}
}
 [2012] Dimitris Bolis, Andras Jakab, Orcun Goksel, and Gabor Szekely:
"On exploiting the connectomics for thalamic nuclei localization: application of pattern recognition techniques",
In European Society for Magnetic Resonance in Medicine and Biology (ESMRMB), Lisbon, Portugal, Oct 2012.
poster
[2012] Dimitris Bolis, Andras Jakab, Orcun Goksel, and Gabor Szekely:
"On exploiting the connectomics for thalamic nuclei localization: application of pattern recognition techniques",
In European Society for Magnetic Resonance in Medicine and Biology (ESMRMB), Lisbon, Portugal, Oct 2012.
poster
@inproceedings{Bolis_exploiting_12a,
author = {Dimitris Bolis and Andras Jakab and Orcun Goksel and Gabor Szekely},
title = {On exploiting the connectomics for thalamic nuclei localization: application of pattern recognition techniques},
booktitle = {European Society for Magnetic Resonance in Medicine and Biology (ESMRMB)},
year = {2012}
}
 [2012] Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Semi-supervised Segmentation Using Multiple Segmentation Hypotheses from a Single Atlas",
In MICCAI W Medical Computer Vision (MCV), pp. 29-37, Nice, France, Oct 2012.
podium
[2012] Tobias Gass, Gabor Szekely, and Orcun Goksel:
"Semi-supervised Segmentation Using Multiple Segmentation Hypotheses from a Single Atlas",
In MICCAI W Medical Computer Vision (MCV), pp. 29-37, Nice, France, Oct 2012.
podium
* Received one of 3 best paper prizes
@inproceedings{Gass_semi-supervised_12,
author = {Tobias Gass and Gabor Szekely and Orcun Goksel},
title = {Semi-supervised Segmentation Using Multiple Segmentation Hypotheses from a Single Atlas},
booktitle = {MICCAI W Medical Computer Vision (MCV)},
year = {2012},
pages = {29-37},
doi = {10.1007/978-3-642-36620-8_4}
}
 [2012] Dimitris Bolis, András Jakab, Orccun Göksel, and Gábor Székely:
"On Exploiting Connectomics for Thalamic Nuclei Localization: A Supervised Learning Approach",
In Int Conf on Machine Learning (ICML) Workshop on Statistics, Machine Learning and Neuroscience, Edinburgh, Scotland, UK, Jul 2012.
[2012] Dimitris Bolis, András Jakab, Orccun Göksel, and Gábor Székely:
"On Exploiting Connectomics for Thalamic Nuclei Localization: A Supervised Learning Approach",
In Int Conf on Machine Learning (ICML) Workshop on Statistics, Machine Learning and Neuroscience, Edinburgh, Scotland, UK, Jul 2012.
@inproceedings{Bolis_exploiting_12,
author = {Dimitris Bolis and Andr\'as Jakab and Or\ccun G\"oksel and G\'abor Sz\'ekely},
title = {On Exploiting Connectomics for Thalamic Nuclei Localization: A Supervised Learning Approach},
booktitle = {Int Conf on Machine Learning (ICML) Workshop on Statistics, Machine Learning and Neuroscience},
year = {2012},
url = {https://sites.google.com/site/stamlins/proceedings}
}
 [2012] Dimitris Bolis, Andras Jakab, Orcun Goksel, and Gabor Szekely:
"Application of pattern recognition techniques to locate the Vim thalamic nucleus based on thalamocortical tractography",
In European Congress on Radiology (ECR), pp. C-1991, Vienna, Austria, Mar 2012.
poster
[2012] Dimitris Bolis, Andras Jakab, Orcun Goksel, and Gabor Szekely:
"Application of pattern recognition techniques to locate the Vim thalamic nucleus based on thalamocortical tractography",
In European Congress on Radiology (ECR), pp. C-1991, Vienna, Austria, Mar 2012.
poster
@inproceedings{Bolis_application_12,
author = {Dimitris Bolis and Andras Jakab and Orcun Goksel and Gabor Szekely},
title = {Application of pattern recognition techniques to locate the Vim thalamic nucleus based on thalamocortical tractography},
booktitle = {European Congress on Radiology (ECR)},
year = {2012},
pages = {C-1991}
}
 [2012] Farheen Taquee, Orcun Goksel, S. Sara Mahdavi, Mira Keyes, W. James Morris, Ingrid Spadinger, and Septimiu Salcudean:
"Deformable Prostate Registration from MR and TRUS Images Using Surface Error Driven FEM models",
In SPIE Medical Imaging, San Diego, CA, USA, Feb 2012.
[2012] Farheen Taquee, Orcun Goksel, S. Sara Mahdavi, Mira Keyes, W. James Morris, Ingrid Spadinger, and Septimiu Salcudean:
"Deformable Prostate Registration from MR and TRUS Images Using Surface Error Driven FEM models",
In SPIE Medical Imaging, San Diego, CA, USA, Feb 2012.
@inproceedings{Taquee_deformable_12,
author = {Farheen Taquee and Orcun Goksel and S. Sara Mahdavi and Mira Keyes and W. James Morris and Ingrid Spadinger and Septimiu Salcudean},
title = {Deformable Prostate Registration from MR and TRUS Images Using Surface Error Driven FEM models},
booktitle = {SPIE Medical Imaging},
year = {2012},
doi = {10.1117/12.911688}
}
 [2011] Orcun Goksel, Kirill Sapchuk, and Septimiu E. Salcudean:
"Haptic simulation of needle and probe interaction with tissue for prostate brachytherapy training",
In IEEE World Haptics Conference (WHC), pp. 7-12, Jun 2011.
podium
[2011] Orcun Goksel, Kirill Sapchuk, and Septimiu E. Salcudean:
"Haptic simulation of needle and probe interaction with tissue for prostate brachytherapy training",
In IEEE World Haptics Conference (WHC), pp. 7-12, Jun 2011.
podium
@inproceedings{Goksel_haptic_11n,
author = {Orcun Goksel and Kirill Sapchuk and Septimiu E. Salcudean},
title = {Haptic simulation of needle and probe interaction with tissue for prostate brachytherapy training},
booktitle = {IEEE World Haptics Conference (WHC)},
year = {2011},
pages = {7-12},
doi = {10.1109/WHC.2011.5945453}
}
 [2010] Orcun Goksel and Septimiu E. Salcudean:
"FEM Simulation of Harmonic Tissue Excitation for Prostate Elastography",
In Int Tissue Elasticity Conf (ITEC), pp. 93, Salt Lake City, UT, USA, Oct 2010.
podium
[2010] Orcun Goksel and Septimiu E. Salcudean:
"FEM Simulation of Harmonic Tissue Excitation for Prostate Elastography",
In Int Tissue Elasticity Conf (ITEC), pp. 93, Salt Lake City, UT, USA, Oct 2010.
podium
@inproceedings{Goksel_fem_10,
author = {Orcun Goksel and Septimiu E. Salcudean},
title = {FEM Simulation of Harmonic Tissue Excitation for Prostate Elastography},
booktitle = {Int Tissue Elasticity Conf (ITEC)},
year = {2010},
pages = {93}
}
 [2010] Hani Eskandari, Orcun Goksel, Septimiu E. Salcudean, and Robert Rohling:
"Bandpass Sampling of High Frequency Tissue Motion",
In Int Tissue Elasticity Conf (ITEC), pp. 71, Salt Lake City, UT, USA, Oct 2010.
podium
[2010] Hani Eskandari, Orcun Goksel, Septimiu E. Salcudean, and Robert Rohling:
"Bandpass Sampling of High Frequency Tissue Motion",
In Int Tissue Elasticity Conf (ITEC), pp. 71, Salt Lake City, UT, USA, Oct 2010.
podium
@inproceedings{Eskandari_bandpass_10,
author = {Hani Eskandari and Orcun Goksel and Septimiu E. Salcudean and Robert Rohling},
title = {Bandpass Sampling of High Frequency Tissue Motion},
booktitle = {Int Tissue Elasticity Conf (ITEC)},
year = {2010},
pages = {71}
}
 [2010] Orcun Goksel, Hani Eskandari, and Septimiu E. Salcudean:
"Mesh Adaptation for Improving Inverse-Problem Reconstruction",
In Int Tissue Elasticity Conf (ITEC), pp. 96, Salt Lake City, UT, USA, Oct 2010.
podium
[2010] Orcun Goksel, Hani Eskandari, and Septimiu E. Salcudean:
"Mesh Adaptation for Improving Inverse-Problem Reconstruction",
In Int Tissue Elasticity Conf (ITEC), pp. 96, Salt Lake City, UT, USA, Oct 2010.
podium
@inproceedings{Goksel_mesh_10,
author = {Orcun Goksel and Hani Eskandari and Septimiu E. Salcudean},
title = {Mesh Adaptation for Improving Inverse-Problem Reconstruction},
booktitle = {Int Tissue Elasticity Conf (ITEC)},
year = {2010},
pages = {96}
}
 [2010] Amir Haddadi, Orcun Goksel, Septimiu E. Salcudean, and Keyvan Hashtrudi-Zaad:
"On the Controllability of Dynamic Model-Based Needle Insertion in Soft Tissue",
In IEEE Eng Medicine and Biology Conf (EMBC), pp. 2287-2291, Buenos Aires, Argentina, Sep 2010.
[2010] Amir Haddadi, Orcun Goksel, Septimiu E. Salcudean, and Keyvan Hashtrudi-Zaad:
"On the Controllability of Dynamic Model-Based Needle Insertion in Soft Tissue",
In IEEE Eng Medicine and Biology Conf (EMBC), pp. 2287-2291, Buenos Aires, Argentina, Sep 2010.
has been an active topic of research in the past decade. Although
dynamic feedback control of needle insertion systems is expected
to provide more accurate target tracking, it has received little
attention due to the fact that most available models for needle-tissue
interaction do not incorporate the dynamics of motions. In this paper,
we study the controllability of rigid or flexible needles inside
soft tissues using mechanical-based dynamic models. The results have
significant implications on the design of suitable feedback controllers
for different types of needle insertion systems.
@inproceedings{Haddadi_controllability_10,
author = {Amir Haddadi and Orcun Goksel and Septimiu E. Salcudean and Keyvan Hashtrudi-Zaad},
title = {On the Controllability of Dynamic Model-Based Needle Insertion in Soft Tissue},
booktitle = {IEEE Eng Medicine and Biology Conf (EMBC)},
year = {2010},
pages = {2287-2291},
doi = {10.1109/IEMBS.2010.5627676}
}
 [2010] Orcun Goksel and Septimiu E. Salcudean:
"Haptic Simulator for Prostate Brachytherapy with Simulated Ultrasound",
In Int Symp Biomedical Simulation (ISBMS), pp. 150-9, Phoenix, AZ, USA, Jan 2010.
podium
[2010] Orcun Goksel and Septimiu E. Salcudean:
"Haptic Simulator for Prostate Brachytherapy with Simulated Ultrasound",
In Int Symp Biomedical Simulation (ISBMS), pp. 150-9, Phoenix, AZ, USA, Jan 2010.
podium
@inproceedings{Goksel_haptic_10,
author = {Orcun Goksel and Septimiu E. Salcudean},
title = {Haptic Simulator for Prostate Brachytherapy with Simulated Ultrasound},
booktitle = {Int Symp Biomedical Simulation (ISBMS)},
year = {2010},
pages = {150-9},
doi = {10.1007/978-3-642-11615-5_16}
}
 [2009] Reza Zahiri-Azar, Orcun Goksel, T.S. Yao, Ehsan Dehghan, Joseph Yan, and Septimiu E. Salcudean:
"Multi-Dimensional Sub-Sample Motion Estimation: Initial Results",
In IEEE Int Ultrasonics Symposium (IUS), Rome, Italy, Sep 2009.
podium
[2009] Reza Zahiri-Azar, Orcun Goksel, T.S. Yao, Ehsan Dehghan, Joseph Yan, and Septimiu E. Salcudean:
"Multi-Dimensional Sub-Sample Motion Estimation: Initial Results",
In IEEE Int Ultrasonics Symposium (IUS), Rome, Italy, Sep 2009.
podium
@inproceedings{Zahiri-Azar_multi-dimensional_09,
author = {Reza Zahiri-Azar and Orcun Goksel and T.S. Yao and Ehsan Dehghan and Joseph Yan and Septimiu E. Salcudean},
title = {Multi-Dimensional Sub-Sample Motion Estimation: Initial Results},
booktitle = {IEEE Int Ultrasonics Symposium (IUS)},
year = {2009},
doi = {10.1109/ULTSYM.2009.0595}
}
 [2009] Orcun Goksel and Septimiu E. Salcudean:
"Automatic Prostate Segmentation from Transrectal Ultrasound Elastography Images Using Geometric Active Contours",
In Int Tissue Elasticity Conf (ITEC), pp. 34, Vlissingen, Netherlands, Sep 2009.
podium
[2009] Orcun Goksel and Septimiu E. Salcudean:
"Automatic Prostate Segmentation from Transrectal Ultrasound Elastography Images Using Geometric Active Contours",
In Int Tissue Elasticity Conf (ITEC), pp. 34, Vlissingen, Netherlands, Sep 2009.
podium
* One of 8 Student Best Paper Finalists
@inproceedings{Goksel_automatic_09,
author = {Orcun Goksel and Septimiu E. Salcudean},
title = {Automatic Prostate Segmentation from Transrectal Ultrasound Elastography Images Using Geometric Active Contours},
booktitle = {Int Tissue Elasticity Conf (ITEC)},
year = {2009},
pages = {34}
}
 [2009] Reza Zahiri-Azar, Orcun Goksel, and Septimiu E. Salcudean:
"Application of 2D Polynomial Fitting to Beam Steering for Motion Estimation with Sub-Sample Accuracy",
In Int Tissue Elasticity Conf (ITEC), pp. 124, Vlissingen, Netherlands, Sep 2009.
podium
[2009] Reza Zahiri-Azar, Orcun Goksel, and Septimiu E. Salcudean:
"Application of 2D Polynomial Fitting to Beam Steering for Motion Estimation with Sub-Sample Accuracy",
In Int Tissue Elasticity Conf (ITEC), pp. 124, Vlissingen, Netherlands, Sep 2009.
podium
@inproceedings{Zahiri-Azar_application_09,
author = {Reza Zahiri-Azar and Orcun Goksel and Septimiu E. Salcudean},
title = {Application of 2D Polynomial Fitting to Beam Steering for Motion Estimation with Sub-Sample Accuracy},
booktitle = {Int Tissue Elasticity Conf (ITEC)},
year = {2009},
pages = {124}
}
 [2009] Reza Zahiri-Azar, Orcun Goksel, and Septimiu E. Salcudean:
"Methods for the Estimation of the Sub-Sample Motion Using Digitized Ultrasound Echo Signals in Three Dimensions",
In Int Tissue Elasticity Conf (ITEC), pp. 88, Vlissingen, Netherlands, Sep 2009.
podium
[2009] Reza Zahiri-Azar, Orcun Goksel, and Septimiu E. Salcudean:
"Methods for the Estimation of the Sub-Sample Motion Using Digitized Ultrasound Echo Signals in Three Dimensions",
In Int Tissue Elasticity Conf (ITEC), pp. 88, Vlissingen, Netherlands, Sep 2009.
podium
@inproceedings{Zahiri-Azar_methods_09,
author = {Reza Zahiri-Azar and Orcun Goksel and Septimiu E. Salcudean},
title = {Methods for the Estimation of the Sub-Sample Motion Using Digitized Ultrasound Echo Signals in Three Dimensions},
booktitle = {Int Tissue Elasticity Conf (ITEC)},
year = {2009},
pages = {88}
}
 [2009] Orcun Goksel and Septimiu E. Salcudean:
"High-Quality Model Generation for Finite Element Simulation of Tissue Deformation",
In MICCAI, pp. 248-256, London, UK, Sep 2009.
poster
[2009] Orcun Goksel and Septimiu E. Salcudean:
"High-Quality Model Generation for Finite Element Simulation of Tissue Deformation",
In MICCAI, pp. 248-256, London, UK, Sep 2009.
poster
* Received one of 15 Hamlyn Robotics Travel Grants
@inproceedings{Goksel_high-quality_09,
author = {Orcun Goksel and Septimiu E. Salcudean},
title = {High-Quality Model Generation for Finite Element Simulation of Tissue Deformation},
booktitle = {MICCAI},
year = {2009},
pages = {248-256},
doi = {10.1007/978-3-642-04271-3_31}
}
 [2009] Sara Mahdavi, Orcun Goksel, and Septimiu E. Salcudean:
"3D Prostate Segmentation in Ultrasound Images Based on Tapered and Deformed Ellipsoids",
In MICCAI, pp. 960-7, London, UK, Sep 2009.
poster
[2009] Sara Mahdavi, Orcun Goksel, and Septimiu E. Salcudean:
"3D Prostate Segmentation in Ultrasound Images Based on Tapered and Deformed Ellipsoids",
In MICCAI, pp. 960-7, London, UK, Sep 2009.
poster
images is required for radiation treatment of prostate cancer. Manual
segmentation is a time-consuming task, the results of which are dependent
on image quality and physicians' experience. This paper introduces
a semi-automatic 3D method based on super-ellipsoidal shapes. It
produces a 3D segmentation in less than 15 seconds using a warped,
tapered ellipsoid fit to the prostate. A study of patient images
shows good performance and repeatability. This method is currently
in clinical use at the Vancouver Cancer Center where it has become
the standard segmentation procedure for low dose-rate brachytherapy
treatment.
@inproceedings{Mahdavi_3d_09,
author = {Sara Mahdavi and Orcun Goksel and Septimiu E. Salcudean},
title = {3D Prostate Segmentation in Ultrasound Images Based on Tapered and Deformed Ellipsoids},
booktitle = {MICCAI},
year = {2009},
pages = {960-7},
doi = {10.1007/978-3-642-04271-3_116}
}
 [2008] Reza Zahiri-Azar, Orcun Goksel, T.S. Yao, Ehsan Dehghan, Joseph Yan, and Septimiu E. Salcudean:
"Methods for the Estimation of Sub-sample Motion of Digitized Ultrasound Echo Signals in 2D",
In IEEE Eng Medicine and Biology Conf (EMBC), pp. 5581-4, Vancouver, BC, Canada, Aug 2008.
podium
[2008] Reza Zahiri-Azar, Orcun Goksel, T.S. Yao, Ehsan Dehghan, Joseph Yan, and Septimiu E. Salcudean:
"Methods for the Estimation of Sub-sample Motion of Digitized Ultrasound Echo Signals in 2D",
In IEEE Eng Medicine and Biology Conf (EMBC), pp. 5581-4, Vancouver, BC, Canada, Aug 2008.
podium
@inproceedings{Zahiri-Azar_methods_08,
author = {Reza Zahiri-Azar and Orcun Goksel and T.S. Yao and Ehsan Dehghan and Joseph Yan and Septimiu E. Salcudean},
title = {Methods for the Estimation of Sub-sample Motion of Digitized Ultrasound Echo Signals in 2D},
booktitle = {IEEE Eng Medicine and Biology Conf (EMBC)},
year = {2008},
pages = {5581-4},
doi = {10.1109/IEMBS.2008.4650479}
}
 [2007] Reza Zahiri-Azar, Orcun Goksel, and Septimiu E. Salcudean:
"Real-Time Tissue Deformation Visualization",
In Int Tissue Elasticity Conf (ITEC), pp. 117, Santa Fe, New Mexico, USA, Nov 2007.
podium
[2007] Reza Zahiri-Azar, Orcun Goksel, and Septimiu E. Salcudean:
"Real-Time Tissue Deformation Visualization",
In Int Tissue Elasticity Conf (ITEC), pp. 117, Santa Fe, New Mexico, USA, Nov 2007.
podium
@inproceedings{Zahiri-Azar_real-time_07,
author = {Reza Zahiri-Azar and Orcun Goksel and Septimiu E. Salcudean},
title = {Real-Time Tissue Deformation Visualization},
booktitle = {Int Tissue Elasticity Conf (ITEC)},
year = {2007},
pages = {117}
}
 [2007] Orcun Goksel and Septimiu E. Salcudean:
"Real-time Synthesis of Image Slices in Deformed Tissue from Nominal Volume Images",
In MICCAI, pp. 401-8, Brisbane, QLD, Australia, Oct 2007.
poster
[2007] Orcun Goksel and Septimiu E. Salcudean:
"Real-time Synthesis of Image Slices in Deformed Tissue from Nominal Volume Images",
In MICCAI, pp. 401-8, Brisbane, QLD, Australia, Oct 2007.
poster
@inproceedings{Goksel_real-time_07,
author = {Orcun Goksel and Septimiu E. Salcudean},
title = {Real-time Synthesis of Image Slices in Deformed Tissue from Nominal Volume Images},
booktitle = {MICCAI},
year = {2007},
pages = {401-8},
doi = {10.1007/978-3-540-75757-3_49}
}
 [2007] Orcun Goksel and Septimiu E. Salcudean:
"Fast B-Mode Ultrasound Image Simulation of Deformed Tissue",
In IEEE Eng Medicine and Biology Conf (EMBC), pp. 87-90, Lyon, France, Aug 2007.
podium
[2007] Orcun Goksel and Septimiu E. Salcudean:
"Fast B-Mode Ultrasound Image Simulation of Deformed Tissue",
In IEEE Eng Medicine and Biology Conf (EMBC), pp. 87-90, Lyon, France, Aug 2007.
podium
@inproceedings{Goksel_fast_07,
author = {Orcun Goksel and Septimiu E. Salcudean},
title = {Fast B-Mode Ultrasound Image Simulation of Deformed Tissue},
booktitle = {IEEE Eng Medicine and Biology Conf (EMBC)},
year = {2007},
pages = {87-90},
doi = {10.1109/IEMBS.2007.4352229}
}
 [2007] Orcun Goksel, Reza Zahiri-Azar, and Septimiu E. Salcudean:
"Simulation of Ultrasound Radio-Frequency Signals in Deformed Tissue for Validation of 2D Motion Estimation with Sub-Sample Accuracy",
In IEEE Eng Medicine and Biology Conf (EMBC), pp. 2159-62, Lyon, France, Aug 2007.
poster
[2007] Orcun Goksel, Reza Zahiri-Azar, and Septimiu E. Salcudean:
"Simulation of Ultrasound Radio-Frequency Signals in Deformed Tissue for Validation of 2D Motion Estimation with Sub-Sample Accuracy",
In IEEE Eng Medicine and Biology Conf (EMBC), pp. 2159-62, Lyon, France, Aug 2007.
poster
@inproceedings{Goksel_simulation_07,
author = {Orcun Goksel and Reza Zahiri-Azar and Septimiu E. Salcudean},
title = {Simulation of Ultrasound Radio-Frequency Signals in Deformed Tissue for Validation of 2D Motion Estimation with Sub-Sample Accuracy},
booktitle = {IEEE Eng Medicine and Biology Conf (EMBC)},
year = {2007},
pages = {2159-62},
doi = {10.1109/IEMBS.2007.4352750}
}
 [2006] Ehsan Dehghan, Orcun Goksel, and Septimiu E. Salcudean:
"A Comparison of Needle Bending Models",
In MICCAI, pp. 305-312, Copenhagen, Denmark, Oct 2006.
poster
[2006] Ehsan Dehghan, Orcun Goksel, and Septimiu E. Salcudean:
"A Comparison of Needle Bending Models",
In MICCAI, pp. 305-312, Copenhagen, Denmark, Oct 2006.
poster
needle insertion simulation and path planning. In this paper, three
models are compared in terms of accuracy in simulating the bending
of a prostate brachytherapy needle. The first two utilize the finite
element method, one using geometric non-linearity and triangular
plane elements, the other using non-linear beam elements. The third
model uses angular springs to model cantilever deflection. The simulations
are compared with the experimental bent needle configurations. The
models are assessed in terms of geometric conformity using independently
identified and pre-identified model parameters. The results show
that the angular spring model, which is also the simplest, simulates
the needle more accurately than the others.
@inproceedings{Dehghan_comparison_06,
author = {Ehsan Dehghan and Orcun Goksel and Septimiu E. Salcudean},
title = {A Comparison of Needle Bending Models},
booktitle = {MICCAI},
year = {2006},
pages = {305-312},
doi = {10.1007/11866565_38}
}
 [2006] Orcun Goksel, Septimiu E. Salcudean, and Robert Rohling:
"Image Synthesis of Deformed Tissue with Application to Ultrasound for Prostate Brachytherapy",
In Canadian Medical and Biological Engineering Conf (CMBEC), Vancouver, BC, Canada, Jun 2006.
podium, national
[2006] Orcun Goksel, Septimiu E. Salcudean, and Robert Rohling:
"Image Synthesis of Deformed Tissue with Application to Ultrasound for Prostate Brachytherapy",
In Canadian Medical and Biological Engineering Conf (CMBEC), Vancouver, BC, Canada, Jun 2006.
podium, national
* First place in the Student Paper Competition
@inproceedings{Goksel_image_06,
author = {Orcun Goksel and Septimiu E. Salcudean and Robert Rohling},
title = {Image Synthesis of Deformed Tissue with Application to Ultrasound for Prostate Brachytherapy},
booktitle = {Canadian Medical and Biological Engineering Conf (CMBEC)},
year = {2006}
}
 [2005] Orcun Goksel, Simon P. DiMaio, Septimiu E. Salcudean, Robert Rohling, and James Morris:
"3D Needle-Tissue Interaction Simulation for Prostate Brachytherapy",
In MICCAI, pp. 827-834, Palm Springs, CA, USA, Oct 2005.
podium
[2005] Orcun Goksel, Simon P. DiMaio, Septimiu E. Salcudean, Robert Rohling, and James Morris:
"3D Needle-Tissue Interaction Simulation for Prostate Brachytherapy",
In MICCAI, pp. 827-834, Palm Springs, CA, USA, Oct 2005.
podium
@inproceedings{Goksel_3d_05,
author = {Orcun Goksel and Simon P. DiMaio and Septimiu E. Salcudean and Robert Rohling and James Morris},
title = {3D Needle-Tissue Interaction Simulation for Prostate Brachytherapy},
booktitle = {MICCAI},
year = {2005},
pages = {827-834},
doi = {10.1007/11566465_102}
}
 [2005] Orcun Goksel, Septimiu E. Salcudean, and Robert Rohling:
"3D Needle-Tissue Interaction Simulation for Prostate Brachytherapy",
In Annual Canadian Conference on Intelligent Systems (IS 2005), 2005.
[2005] Orcun Goksel, Septimiu E. Salcudean, and Robert Rohling:
"3D Needle-Tissue Interaction Simulation for Prostate Brachytherapy",
In Annual Canadian Conference on Intelligent Systems (IS 2005), 2005.
* Received the Best Poster Award
@inproceedings{Goksel_3Dis_05,
author = {Orcun Goksel and Septimiu E. Salcudean and Robert Rohling},
title = {3D Needle-Tissue Interaction Simulation for Prostate Brachytherapy},
booktitle = {Annual Canadian Conference on Intelligent Systems (IS 2005)},
year = {2005}
}
 [2004] Orcun Goksel, Septimiu E. Salcudean, and Robert Rohling:
"Towards a Prostate Brachytherapy Haptic Simulator",
In BC Advanced Systems Institute (ASI) Exchange, 2004.
poster
[2004] Orcun Goksel, Septimiu E. Salcudean, and Robert Rohling:
"Towards a Prostate Brachytherapy Haptic Simulator",
In BC Advanced Systems Institute (ASI) Exchange, 2004.
poster
* Received the ASI Innovation Award and Communication Awards
@inproceedings{Goksel_towards_04,
author = {Orcun Goksel and Septimiu E. Salcudean and Robert Rohling},
title = {Towards a Prostate Brachytherapy Haptic Simulator},
booktitle = {BC Advanced Systems Institute (ASI) Exchange},
year = {2004}
}
Monographs:
 [2009] Orcun Goksel:
"Meshing and Rendering of Patient-Specific Deformation Models With Application to Needle Insertion Simulation",
University of British ColumbiaUniversity of British Columbia, 2009.
[2009] Orcun Goksel:
"Meshing and Rendering of Patient-Specific Deformation Models With Application to Needle Insertion Simulation",
University of British ColumbiaUniversity of British Columbia, 2009.
* Western Association of Graduate Schools (WAGS) Innovation in Technology Award for the top-ranked dissertation of the year for western North America
@phdthesis{Goksel_meshing_09,
author = {Orcun Goksel},
title = {Meshing and Rendering of Patient-Specific Deformation Models With Application to Needle Insertion Simulation},
school = {University of British Columbia},
year = {2009},
url = {http://hdl.handle.net/2429/17418},
doi = {10.14288/1.0068717}
}
 [2005] Orcun Goksel:
"Ultrasound Image and 3D Finite Element based Tissue Deformation Simulator for Prostate Brachytherapy",
University of British ColumbiaUniversity of British Columbia, 2005.
[2005] Orcun Goksel:
"Ultrasound Image and 3D Finite Element based Tissue Deformation Simulator for Prostate Brachytherapy",
University of British ColumbiaUniversity of British Columbia, 2005.
@mastersthesis{Goksel_ultrasound_05,
author = {Orcun Goksel},
title = {Ultrasound Image and 3D Finite Element based Tissue Deformation Simulator for Prostate Brachytherapy},
school = {University of British Columbia},
year = {2005},
url = {http://hdl.handle.net/2429/16194},
doi = {10.14288/1.0065481}
}
Non-reviewed Preprints and Technical Reports (Online / ArXiv):
 [2024] Lingkai Zhu, Can Deniz Bezek, and Orcun Goksel:
"FGGP: Fixed-Rate Gradient-First Gradual Pruning",
(arxiv:2411.05500), 2024.
[2024] Lingkai Zhu, Can Deniz Bezek, and Orcun Goksel:
"FGGP: Fixed-Rate Gradient-First Gradual Pruning",
(arxiv:2411.05500), 2024.
@unpublished{Zhu_fggp_24,
author = {Lingkai Zhu and Can Deniz Bezek and Orcun Goksel},
title = {FGGP: Fixed-Rate Gradient-First Gradual Pruning},
year = {2024},
number = {arxiv:2411.05500},
doi = {10.48550/arXiv.2411.05500}
}
 [2024] Alvaro Gomariz, Yusuke Kikuchi, Yun Yvonna Li, Thomas Albrecht, Andreas Maunz, Daniela Ferrara, Huanxiang Lu, and Orcun Goksel:
"Joint semi-supervised and contrastive learning enables zero-shot domain-adaptation and multi-domain segmentation",
(arxiv:2405.05336), 2024.
[2024] Alvaro Gomariz, Yusuke Kikuchi, Yun Yvonna Li, Thomas Albrecht, Andreas Maunz, Daniela Ferrara, Huanxiang Lu, and Orcun Goksel:
"Joint semi-supervised and contrastive learning enables zero-shot domain-adaptation and multi-domain segmentation",
(arxiv:2405.05336), 2024.
@unpublished{Gomariz_joint_24,
author = {Alvaro Gomariz and Yusuke Kikuchi and Yun Yvonna Li and Thomas Albrecht and Andreas Maunz and Daniela Ferrara and Huanxiang Lu and Orcun Goksel},
title = {Joint semi-supervised and contrastive learning enables zero-shot domain-adaptation and multi-domain segmentation},
year = {2024},
number = {arxiv:2405.05336},
doi = {10.48550/arxiv.2211.04238}
}
 [2023] Lin Zhang, Tiziano Portenier, and Orcun Goksel:
"Unpaired Translation from Semantic Label Maps to Images by Leveraging Domain-Specific Simulations",
(arxiv:2302.10698), 2023.
[2023] Lin Zhang, Tiziano Portenier, and Orcun Goksel:
"Unpaired Translation from Semantic Label Maps to Images by Leveraging Domain-Specific Simulations",
(arxiv:2302.10698), 2023.
@unpublished{Zhang_unpaired_23,
author = {Lin Zhang and Tiziano Portenier and Orcun Goksel},
title = {Unpaired Translation from Semantic Label Maps to Images by Leveraging Domain-Specific Simulations},
year = {2023},
number = {arxiv:2302.10698},
doi = {10.48550/ARXIV.2302.10698}
}
 [2022] Alina C. Teuscher, Cyril Statzer, Anita Goyala, Seraina A. Domenig, Ingmar Schoen, Max Hess, Alexander M. Hofer, Andrea Fossati, Viola Vogel, Orcun Goksel, Ruedi Aebersold, and Collin Y. Ewald:
"Mechanotransduction Coordinates Extracellular Matrix Protein Homeostasis Promoting Longevity in C. Elegans",
, 2022.
[2022] Alina C. Teuscher, Cyril Statzer, Anita Goyala, Seraina A. Domenig, Ingmar Schoen, Max Hess, Alexander M. Hofer, Andrea Fossati, Viola Vogel, Orcun Goksel, Ruedi Aebersold, and Collin Y. Ewald:
"Mechanotransduction Coordinates Extracellular Matrix Protein Homeostasis Promoting Longevity in C. Elegans",
, 2022.
@unpublished{Teuscher_mechanotransduction_22,
author = {Alina C Teuscher and Cyril Statzer and Anita Goyala and Seraina A Domenig and Ingmar Schoen and Max Hess and Alexander M. Hofer and Andrea Fossati and Viola Vogel and Orcun Goksel and Ruedi Aebersold and Collin Y. Ewald},
title = {Mechanotransduction Coordinates Extracellular Matrix Protein Homeostasis Promoting Longevity in C. Elegans},
year = {2022},
doi = {10.1101/2022.08.30.505802}
}
 [2021] Hossein Khodadadi, Orcun Goksel, and Sabine Kling:
"Motion Estimation for Optical Coherence Elastography Using Signal Phase and Intensity",
(arxiv:2103.10784), 2021.
[2021] Hossein Khodadadi, Orcun Goksel, and Sabine Kling:
"Motion Estimation for Optical Coherence Elastography Using Signal Phase and Intensity",
(arxiv:2103.10784), 2021.
@unpublished{Khodadadi_motion_21,
author = {Hossein Khodadadi and Orcun Goksel and Sabine Kling},
title = {Motion Estimation for Optical Coherence Elastography Using Signal Phase and Intensity},
year = {2021},
number = {arxiv:2103.10784},
url = {https://arxiv.org/abs/2103.10784}
}
 [2020] Firat Ozdemir, Christine Tanner, and Orcun Goksel:
"Delineating Bone Surfaces in B-Mode Images Constrained by Physics of Ultrasound Propagation",
(arxiv:2001.02001):1-11, Jan 2020.
[2020] Firat Ozdemir, Christine Tanner, and Orcun Goksel:
"Delineating Bone Surfaces in B-Mode Images Constrained by Physics of Ultrasound Propagation",
(arxiv:2001.02001):1-11, Jan 2020.
@unpublished{Ozdemir_delineating_20,
author = {Firat Ozdemir and Christine Tanner and Orcun Goksel},
title = {Delineating Bone Surfaces in B-Mode Images Constrained by Physics of Ultrasound Propagation},
year = {2020},
number = {arxiv:2001.02001},
pages = {1-11},
url = {https://arxiv.org/abs/2001.02001}
}
 [2018] Christine Tanner, Firat Ozdemir, Romy Profanter, Valeriy Vishnevsky, Ender Konukoglu, and Orcun Goksel:
"Generative Adversarial Networks for MR-CT Deformable Image Registration",
(arXiv:1807.07349):1-11, Jul 2018.
[2018] Christine Tanner, Firat Ozdemir, Romy Profanter, Valeriy Vishnevsky, Ender Konukoglu, and Orcun Goksel:
"Generative Adversarial Networks for MR-CT Deformable Image Registration",
(arXiv:1807.07349):1-11, Jul 2018.
@unpublished{Tanner_generative_18,
author = {Christine Tanner and Firat Ozdemir and Romy Profanter and Valeriy Vishnevsky and Ender Konukoglu and Orcun Goksel},
title = {Generative Adversarial Networks for MR-CT Deformable Image Registration},
year = {2018},
number = {arXiv:1807.07349},
pages = {1-11},
url = {https://arxiv.org/abs/1807.07349}
}
 [2016] Ece Ozkan, Gemma Roig, Orcun Goksel, and Xavier Boix:
"Herding Generalizes Diverse M-Best Solutions",
(arXiv:1611.04353):1-8, Nov 2016.
[2016] Ece Ozkan, Gemma Roig, Orcun Goksel, and Xavier Boix:
"Herding Generalizes Diverse M-Best Solutions",
(arXiv:1611.04353):1-8, Nov 2016.
@unpublished{Ozkan_herding_17,
author = {Ece Ozkan and Gemma Roig and Orcun Goksel and Xavier Boix},
title = {Herding Generalizes Diverse M-Best Solutions},
year = {2016},
number = {arXiv:1611.04353},
pages = {1-8},
url = {https://arxiv.org/abs/1611.04353}
}
 [2011] Reza Zahiri Azar, Orcun Goksel, Ehsan Dehghan, and Septimiu E. Salcudean:
"Strain Tensor Visualization Using Glyph Objects: Application to Elastography",
:1-4, Jun 2011.
[2011] Reza Zahiri Azar, Orcun Goksel, Ehsan Dehghan, and Septimiu E. Salcudean:
"Strain Tensor Visualization Using Glyph Objects: Application to Elastography",
:1-4, Jun 2011.
@unpublished{Azar_strain_11,
author = {Reza Zahiri Azar and Orcun Goksel and Ehsan Dehghan and Septimiu E Salcudean},
title = {Strain Tensor Visualization Using Glyph Objects: Application to Elastography},
year = {2011},
pages = {1-4},
doi = {10.13140/RG.2.2.31558.37446}
}




